Identifying Unusual Features in Structure-Based Drug Design
Here we illustrate how Mogul software can be used in structure-based drug design to help identify unusual features in structures and distinguish between crystallographic anomalies and genuinely unusual geometries.
Where Can Mogul Be Used in the Drug Discovery Process?
Mogul provides precise information on preferred molecular geometries within a crystal structure by mining millions of chemically classified bond lengths, angles, torsion angles, and ring conformations in the Cambridge Structural Database (CSD).
Mogul is used to help design and assess new materials, such as drug molecules, and analyse and validate molecular conformations and geometries.
In the specific case of structure-based drug discovery, Mogul is applied:
- To validate ligand structures of interest in the PDB;
- To validate in house protein-ligand structures and identify unusual ring and torsion assignments;
- To filter out poor geometry ligand poses from docking results;
- To identify true ligand strain in a protein-bound ligand that you might be able to alleviate by appropriate ligand design.
Torsions and Rings: Why Are They So Important?
When solving a crystal structure, torsions are often defined solely by fit to electron density and hence unusual torsions are the most common form of unusual geometry.
Identifying an unusual geometry in non-aromatic rings often means that the input model that the user started with is wrong.
In both the cases, Mogul can help identify poor geometries in the structure and discover crucial errors at an early stage.
Errors Occur Even in High Resolution Structures
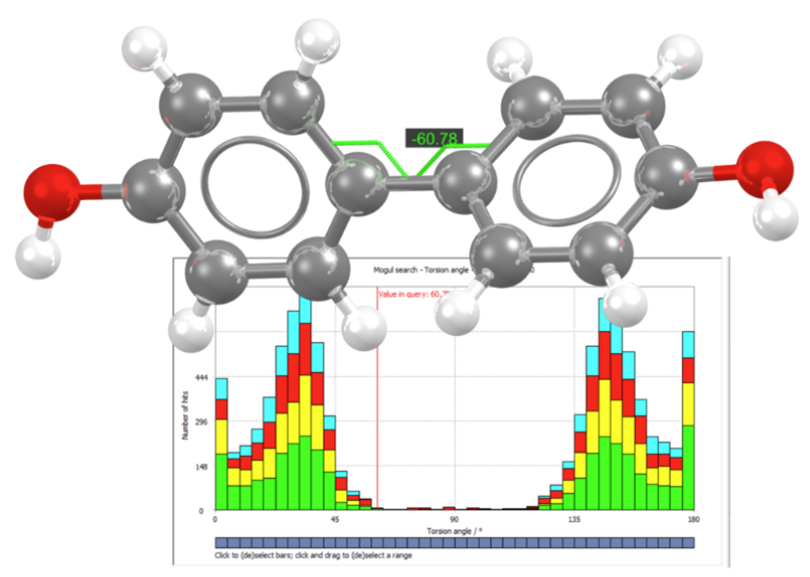
Here we illustrate an example in which the Mogul analysis was performed on the glycosidic ligand in endo-1,3-beta glucanase (2cl2, Figure 1).
This structure is of high resolution, 1.35 Å, and upon visual inspection the flexible ligand seems to fit well to the electron density. However, the Mogul analysis identifies two unusual torsions (listed in orange in Figure 2) corresponding to the two amide bonds in the ligand.
Figure 2 reports the histogram obtained from the Mogul search for one of the two unusual torsions, where it can be seen that the distribution for the selected torsion is expected to be close to 0°, but in the observed structure it is instead close to 180°.
Upon further inspection, it can be concluded that the ligand contains two amide bonds that are set as cis rather than trans, one of which is highlighted in Figure 3. This example shows that an error can occur even in a high-resolution structure, which could have been easily identified using Mogul. To find out more, read the full article here [1].
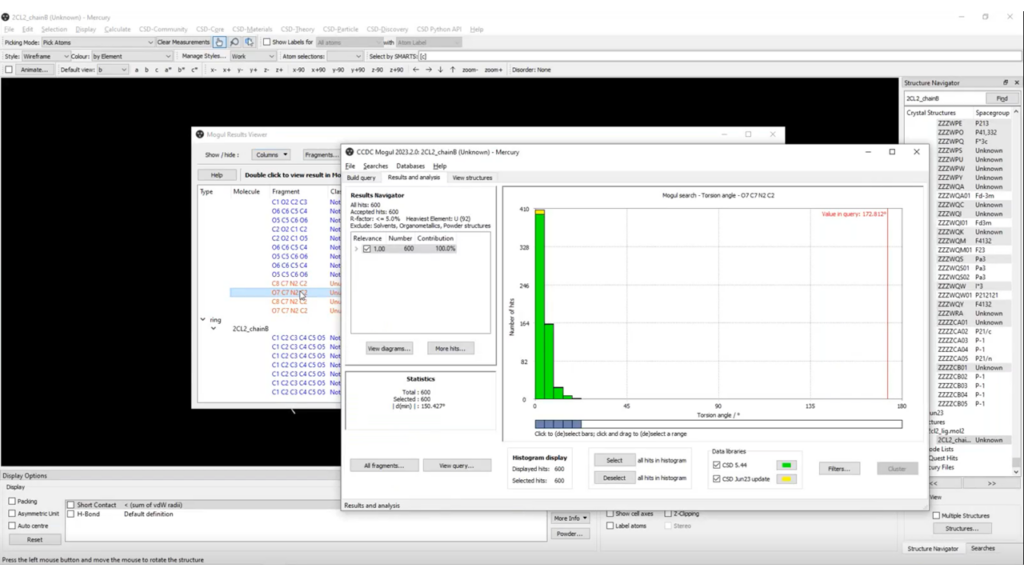
Analysis of Ligand Geometry in Proteins
How often should we expect unusual geometry in drugs bound to proteins? Work published in the Journal of Medicinal Chemistry [2] used Mogul as the tool of choice and examined the ligand geometry in protein structures.
Ligands with at least one rotatable bond, different resolutions, and diverse structures were investigated. A set of small molecule drugs from the CSD was also included as a comparison test set.
The results obtained revealed that for both high- and medium-resolution structures, unusual geometry is rare in crystallographic ligand models that fit the electron density well. By contrast, where fit to electron density is poor, unusual geometry is more common. An example of the latter is the case in which the ligand can adopt multiple binding conformations. In such cases, research still needs to be done to find more robust crystallographic refinement methods.
It can be concluded that where the crystallography is solid, Mogul can be used to identify true unusual geometry within a ligand. Read the full article here [2].
What Can a Drug Designer Do If They Find True Unusual Geometry?
The drug molecules found in the PDB are highly optimised for potency, and perhaps any strain energy on binding is likely to have been minimized during the design process.
This may not always be the case for lead molecules. If you find unusual geometry in your lead molecule, here’s a few solutions:
- Relief of unusual geometry by chemical modification may reduce relievable strain and increase potency;
- Freezing of unusual geometry by (macro)cyclization may reduce relievable strain and increase potency;
- The use of Mogul may help identify a problem in the geometry.
An interesting example published in Bioorganic & Medicinal Chemistry Letters [3] shows the approach that the scientists at Bristol-Myers Squibb used to discover a series of FXIa inhibitors with improved potency.
The team identified that the acrylamide portion in one of their lead structures was sitting in a high energy torsion space and redesigned the molecule replacing the acrylamide with a cyclic carbamate. Cyclization was hence used to remove the unusual geometry and led to a 33x increase in affinity.
Discover more about the importance of torsions in drug design and learn how to perform a conformational analysis in our blog “How Torsional Data Can Inspire Drug Design”.
Conclusions
We demonstrated how Mogul is an excellent tool for identifying crystallographic problems with ligands in protein-ligand crystal structures.
True unusual geometry within a ligand can be easily identified with Mogul when the crystallography is solid, and the use of molecular design to eliminate the unusual geometry may reduce strain and lead to a more potent ligand.
Next Steps
Are you interested in finding out more about Mogul, including its implementation in the industrial research laboratory of Astex Pharmaceuticals, a global developer of small molecule drugs? Watch on demand the webinar “How Rapid Assessment of Molecular Geometries Can Help the Structure-Based Drug Designer”, presented by John Liebeschuetz, Computational Chemist at Astex Pharmaceuticals.
To discuss further and/or request a demo with one of our scientists, please contact us via this form or .
References
[1] Liebeschuetz, J. et al. J Comput Aided Mol Des 26, 169–183 (2012).
[2] Liebeschuetz, J, J. Med. Chem. 64, 7533–7543 (2021).
[3] Clark, C. G. et al. Bioorganic & Medicinal Chemistry Letters 29, 126604 (2019).