CSD in action: Novartis. CSD-validated data helps discover compounds with 50% increase in potency for treatment of SMA
In this article we discuss research carried out by scientists at Novartis using the Cambridge Structural Database (CSD) in the development of potent small-molecule splicing modulators for the treatment of spinal muscular atrophy (SMA). This is part of our series highlighting examples of the Cambridge Crystallographic Data Centre (CCDC) tools in action by scientists around the world.
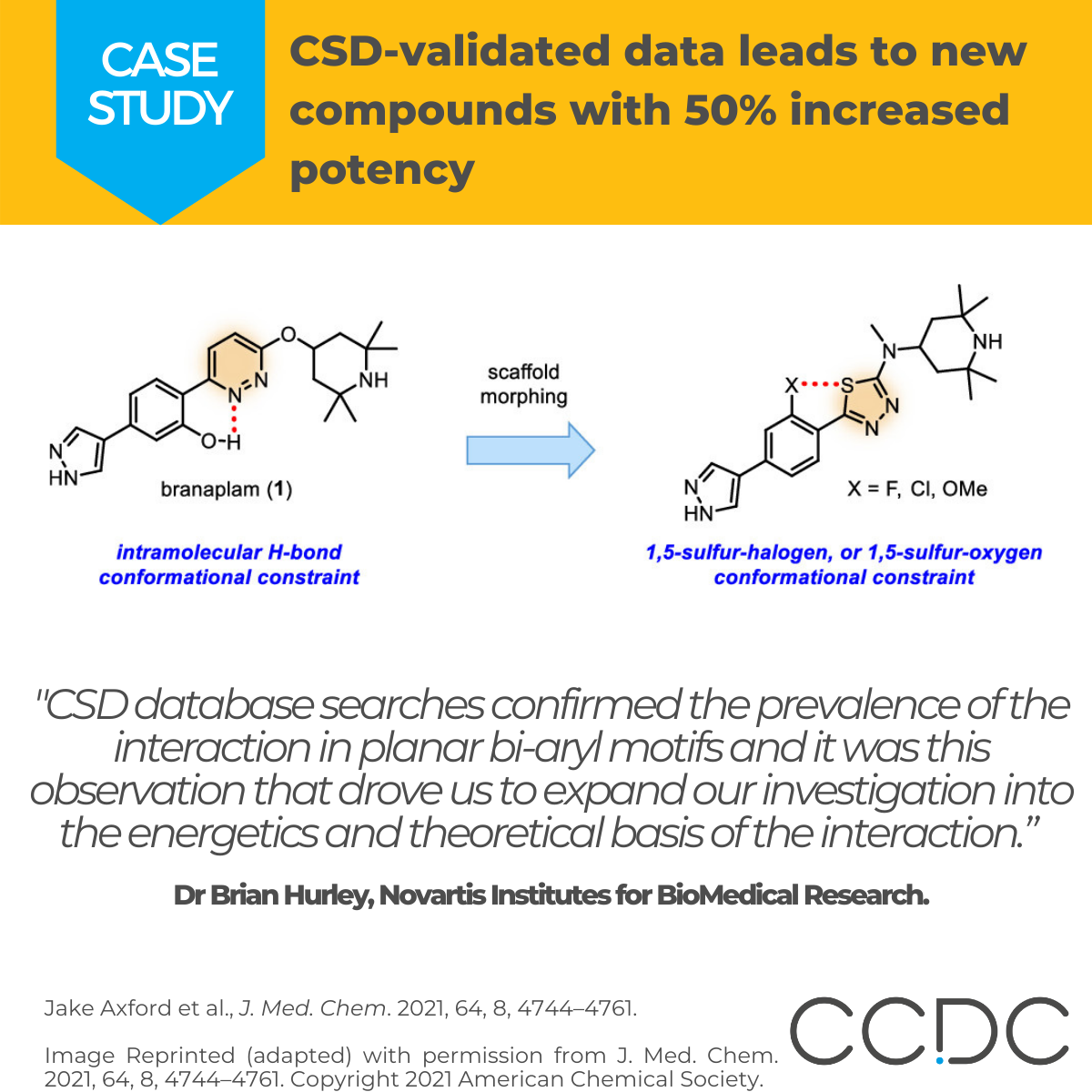
“CSD database searches confirmed the prevalence of the interaction in planar bi-aryl motifs and it was this observation that drove us to expand our investigation into the energetics and theoretical basis of the interaction. It’s clear that the arsenal of intra- and inter-molecular interactions available to medicinal chemists is still evolving, as is our understanding of how to best take advantage of them.”
Dr Brian Hurley, Senior Principal Scientist, Global Discovery Chemistry, Novartis Institutes for BioMedical Research.
Summary
A new class of small molecules with 50% increase in potency for the treatment of SMA has been discovered by researchers from Novartis Institutes for BioMedical Research. The team used CSD-validated data to understand the nature of interactions responsible for conformational constraints in the molecules under investigation.
Why
SMA is a debilitating neuromuscular disease affecting muscle strength that leads to mobility problems. There are four types of SMA that affect people of different ages from babies to adults, and the condition gets worse with time.
SMA is caused by depleted levels of the survival motor neuron (SMN) protein caused by mutations in a gene called SMN1.Neighbouring SMN2 genes can in part compensate for non-functional SMN1 genes by encoding for full and truncated SMN protein. Because of its disease-modifying properties, SMN2 has been targeted for potential SMA therapy, as an alternative way to produce SMN protein.
This work adds to the significant efforts being made in the development of brain-penetrant, and orally bioavailable small-molecule therapeutics able to increase levels of full-length SMN protein by acting as SMN2 splicing modulators.
How did the CSD help in this research?
The authors identified the nearly coplanar conformation of the pyridazine-phenyl biaryl system required for efficacy, and hypothesized that factors favouring planar conformer populations of the biaryl system would result in improved ligand-target binding.Scaffold morphing (changing core structures) revealed a favourable replacement of the pyridazine core with a 1,3,4-thiadiazole ring system that provided additional opportunities for planarizing conformational constraints of the biaryl through intramolecular 1,5-sulfur−oxygen (S···O) or 1,5-sulfur halogen (S···X) noncovalent interactions.
The CSD was then mined to reveal the exact nature of the sulfur—halogen interaction responsible for the conformational constraint. 30+ molecules of interest were at first identified with sulfur—fluorine interactions with a further 6 molecules where the nature of sulfur—chlorine interactions was revealed.
The initial oxygen-based conformational constraints (−OH, −OMe) were then replaced with a fluorine or chlorine resulting in a new 2-halophenyl thiadiazole motif series with potency, pharmacokinetics, and central nervous system distribution sufficient to provide 50% increase in production of full-length SMN protein in a mouse model of SMA.
Whilst intramolecular sulfur-halogen (S···X) interactions have found practical applications in materials science, this work shows that sulfur-halogen chalcogen bonding provide attractive alternatives to intramolecular hydrogen-bonding and S···O interactions in drug design.
Read the full paper: Use of Intramolecular 1,5-Sulfur–Oxygen and 1,5-Sulfur–Halogen Interactions in the Design of N-Methyl-5-aryl-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3,4-thiadiazol-2-amine SMN2 Splicing Modulators, J. Med. Chem. 2021, 64, 8, 4744–4761.