CSD Python API for Ensemble Docking
Today’s blog is based on the webinar ‘CSD Python API for Ensemble Docking’ (access the recording here), and illustrates the basics of ensemble docking, providing an introduction to the CSD Python API toolkit. Watch on demand the full webinar.
Docking and Ensembles – The Basics
Docking is a computational method used to explore the possible binding modes of a substrate to a given receptor, enzyme, or other binding sites. Ensemble refers to a group of objects, or people acting or taken together as a whole.
Ensemble docking is a docking technique in which the binding mode of the substrate is evaluated across multiple receptor structures.
Importance and Challenges of Molecular Docking
Molecular docking is widely recognized as pivotal in drug design and discovery. Its importance derives from the beneficial applications: the identification of bioactive conformations and compounds that bind to specific targets of interest; and efficient virtual screening experiments.
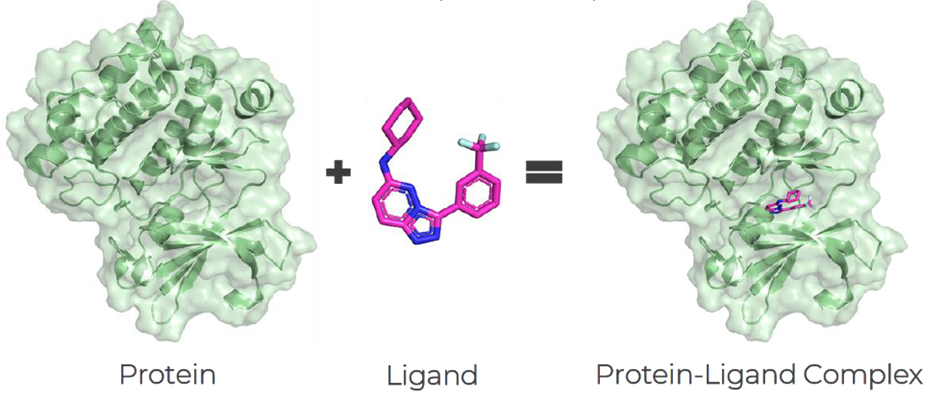
One of the biggest challenges of molecular docking is protein flexibility. Most of the software available today can account for small conformational changes, such as movement of side chains. However, for more large-scale structural rearrangements, such as movements of the backbone of the protein, the task is more difficult.
A possible solution is ensemble docking that accounts for several discrete protein conformations for docking. It is important to include protein structures that capture different conformational states and conformations of the binding site: examples are the inclusion of protein structures which are bound to different ligands, or even unbound structures; structures determined crystallographically, by NMR, or by other methods; and the inclusion of different time snapshots of the molecular dynamic simulation.
Molecular Docking with GOLD
GOLD (‘Genetic Optimization for Ligand Docking’) is the validated, configurable protein–ligand docking software for expert drug discovery. It is part of the CSD-Discovery suite that provides all the tools needed to design and discover new molecules.
With GOLD, researchers can easily search for conformational ensembles. It can be accessed by a user-friendly interface via the software Hermes, or programmatically through the CSD Python API.
Why an API? Why Python?
APIs (Application Programming Interfaces) promote interoperability, modularity, platform independence, innovation, and third-party integration. With the CSD Python API it is possible to access CSD-based data without being bound by the constraints of the CSD graphical user interface, and create workflows which are tailored to the specific needs of the user.
Python is amongst the top programming languages, and has become very popular especially in the scientific community, with documentation, tutorials, and community support. There are several libraries in Python for data manipulation, scientific computing, and machine learning, making Python an excellent choice for rapid prototyping and development.
How to Access the CSD Python API
There are several ways to access the CSD Python API: the 2023.2 CSD software release includes a self-contained, ready to be used Python environment (Miniconda) with the CSD Python API and all its prerequisites. Otherwise, it is possible to install it in an environment of the user’s choice via Conda or pip packages; additionally, it can be also accessed using Jupiter notebooks; finally, for users that are more comfortable using graphical user interface applications, it is possible to find ready to be used scripts that can be run directly form the software Mercury or Hermes.
Generating 3D Molecules and Similarity Search
The following cases represent two simple examples of what can be achieved with CSD Python API.
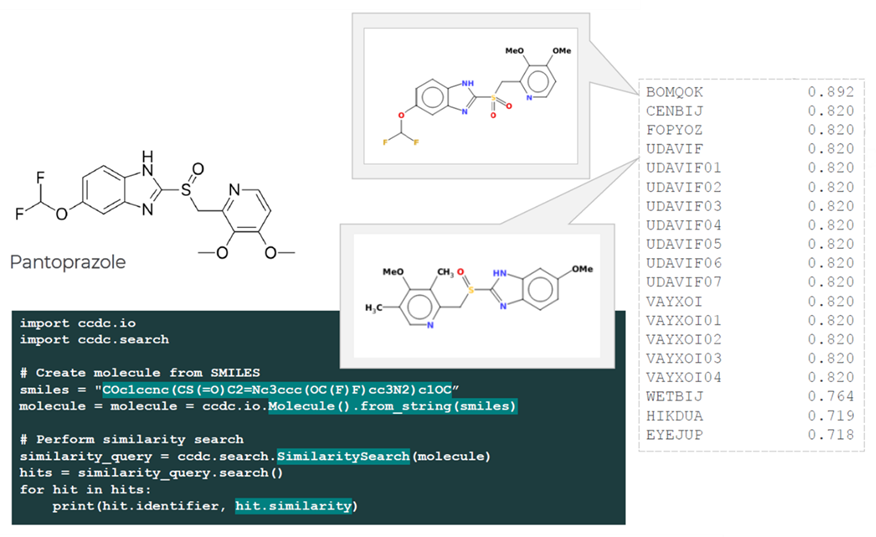
The lines of code that can be seen in Figure 2 generate 3D conformations of a molecule starting just from a SMILES code for that molecule.

In Figure 3, a similarity search was performed scanning the more than 1.2 million structures present in the CSD for molecules which are similar to a certain target.
CSD Python API: Access to Documentation and Case Study
More in-depth searching and chemical analyses can be done with the CSD Python API. A detailed documentation can be accessed here, containing descriptive instructions on how to perform the tasks, and details of the different modules present in the API.
A case study was illustrated in detail during the webinar demo, where the CSD Python API was used to perform ensemble docking on the protein thymidine kinase (access the recording here).
Next Steps
To discuss further and/or request a demo with one of our scientists, please contact us via this form or .