Protein-ligand docking 101 - running a simulation in GOLD
Introduction
Molecular docking applications in drug discovery are varied, including structure-activity studies, lead optimisation, finding potential leads by virtual screening, providing binding hypotheses to facilitate predictions for mutagenesis studies and, also in assisting X-ray and cryogenic electron microscopy (cryo-EM) crystallography in the fitting of substrates and inhibitors to electron density.
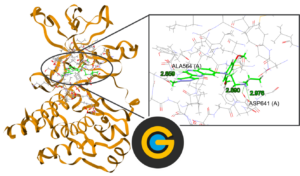
GOLD (Genetic Optimisation for Ligand Docking) is a genetic algorithm for docking flexible ligands into protein binding sites. GOLD has been extensively tested and has shown excellent performance for pose prediction and good results for virtual screening.
Learning Outcomes
- Familiarity of proteins, ligands and functional waters.
- Familiarity of what protein ligand docking is and why you might want to perform it.
- Familiarity with the GOLD interface and a step-by-step guide of the basic functionality.
- Where to get started with your docking simulation.
- The basics of how to run a standard protein-ligand dock with GOLD.
Requirements
- The CSD installed and a current valid CSD licence active. For installation and licences visit the FAQs.
- This module is designed for beginners of the docking software GOLD.
This module takes less than 90 minutes to complete. Those that complete this module and answer the final quiz questions correctly will gain a LinkedIn badge certification to showcase their achievement.
Your CSDU Module
U Watch

This playlist introduces the module, explains what the CSD is, introduces protein-ligand docking and GOLD, and demonstrates how to use GOLD to run protein-docking simulation with a step-by-step tutorial.
If you have completed another CSDU module before, you can start from the second video.
The slides for the introductory video are available here and also an additional deck of slides with step-by-step procedure for the case study shown in the demo is available here.
If YouTube is not available in your country, you can contact us for alternative access to the videos.
U Test

Test what you have learnt in this quick quiz to finish the module, and receive a LinkedIn badge certification to showcase on your LinkedIn profile.
You will need to score 10/10 to receive the certification badge. Do not worry if you don’t pass it at the first attempt: you can take this test multiple times!
It may take up to few hours to receive your certification badge. If you do not receive it within a day, please contact us!
