It’s the World Health Organization’s World Antimicrobial Awareness Week
Every year, the World Health Organization (WHO) celebrates World Antimicrobial Awareness Week (WAAW), which aims to increase awareness of global antimicrobial resistance to help stop the emergence and spread of drug-resistant infections. The WAAW 2021 theme is “Spread Awareness, Stop Resistance.” In this blog, we look at antimicrobial structures in the Cambridge Structural Database (CSD), as well as how computational and experimental chemists leverage the data to develop new therapeutics.
How many structures in the CSD have antimicrobial properties?
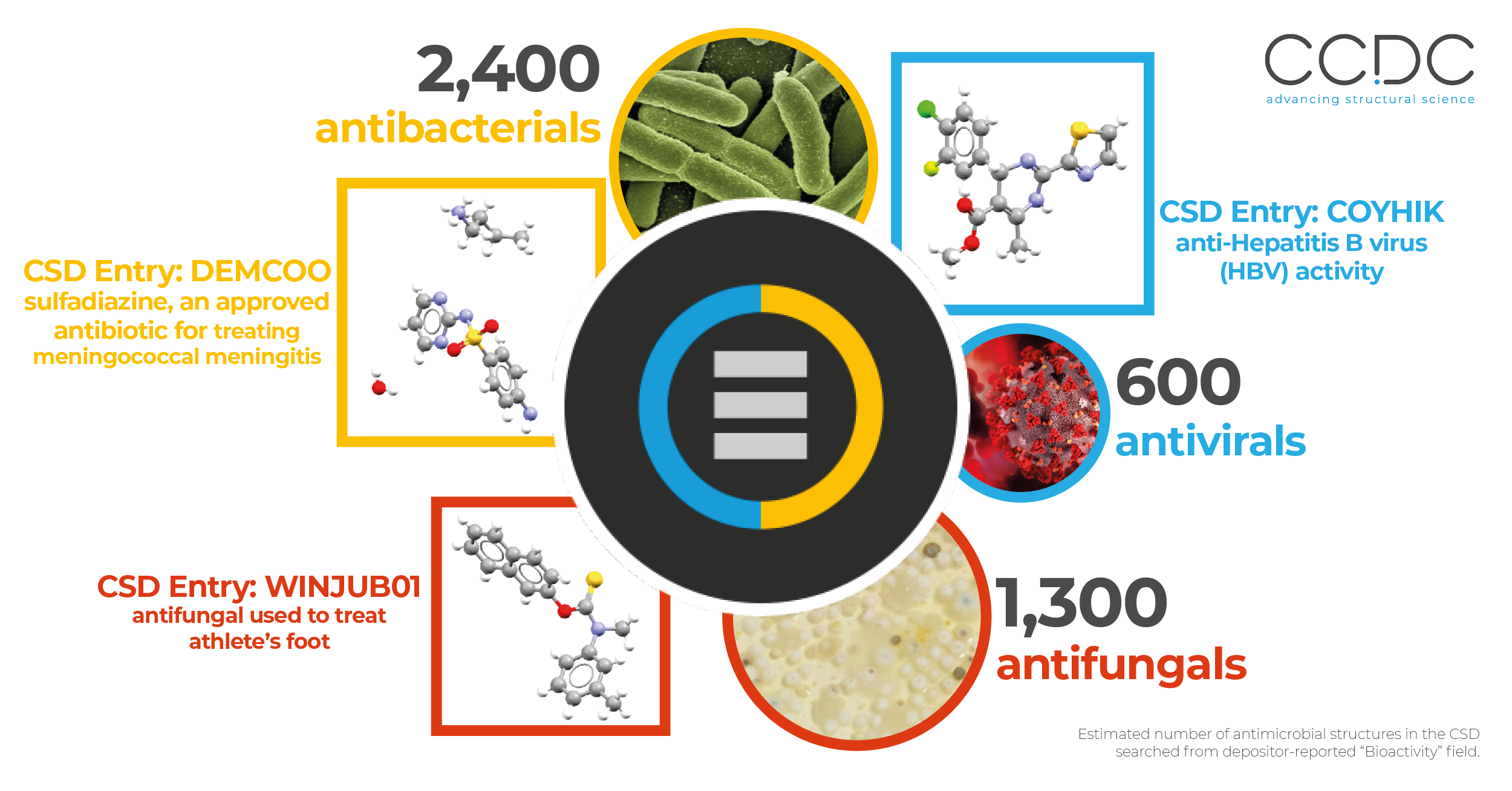
Based on information depositors include with their structures and information that CSD editors can collect from the depositors’ published work, we know the CSD contains at least 5,400 compounds with potential antimicrobial properties. We also know the CSD contains at least:
- 2,400 compounds with potential antibacterial properties.
- 600 compounds with potential antiviral properties.
- 1,300 compounds with potential antifungal properties.
There are also nearly 400 compounds with expected antiprotozoal properties, including many against the plasmodium parasite that causes malaria. This is based on searching for key terms in the CSD’s optional “bioactivity” field.
How can researchers use data in the CSD to develop new antimicrobials?
The amount and quality of the data in the CSD mean it plays a vital role in the development of new antimicrobial therapeutics. Researchers worldwide use the experimentally derived data in the CSD to inform the design of new therapeutics and then de-risk them with computational techniques. This can save valuable time at the experimental phases of the drug discovery pipeline. For example, the data can help answer questions, like:
- What are the preferred molecular geometries of my system and does it have any unusual features?
- Are there “hot spots” that exist on a protein that may provide binding sites to a potential small molecule?
Combining computational and experimental analysis improves the antimicrobial pipeline
Computational approaches that leverage the CSD as a knowledge base, as well as molecular docking approaches that analyze interactions that occur between proteins and compounds of interest, can help focus efforts at the benchtop. According to a recent article in Computational and Structural Biotechnology Journal:
- “[T]he use of computational approaches for initial virtual screening, followed by concurrent experimental and computational analysis has the potential to reduce costs and increase the quality of compounds taken forward towards the developmental pipeline.”
- “Molecular docking is the most widely used computational approach for virtual screening against M. tuberculosis proteins and has resulted in numerous published studies.”
By working together, computational and experimental chemists can more quickly develop new pharmaceuticals and address antibiotic resistance in microbes like Mycobacterium tuberculosis—the causative agent of tuberculosis (TB). According to the World Health Organization’s 2020 Global Tuberculosis Report:
- In 2019, TB contributed to an estimated 1.4 million deaths, with approximately 10 million new infections in the same year.
- Resistance to the current drug-treatment regime is rising, with 3.3% of new TB cases being multi-drug resistant (MDR) and 17.7% of previously treated TB cases being MDR.
CCDC’s protein docking software, GOLD, allows researchers to programmatically search for new potential pharmaceuticals that may bind with important proteins that microorganisms need for metabolism and other vital functions. For example, researchers at CCDC and Durham University used GOLD to identify novel scaffolds of EthR inhibitors, which then went on to biophysical assay tests. EthR is a member of the TetR family involved in M. tuberculosis’s resistance to ethionamide. Ethionamide is an important proto-drug for the treatment of TB that limits processes essential to maintaining the pathogen’s cell wall. They published their findings in the journal, Organic & Biomolecular Chemistry.
Molecular docking can play a key role in finding new antimicrobials of all kinds. For example, in a paper in Scientific Reports, researchers used GOLD to screen a library of 6,900 approved drugs to see if any inhibit the main protease of SARS-CoV-2 (COVID-19). Again this guided experimental work, with the best few tested by activity assay for further characterization.
Read more
Learn how you can access the data stored in the CSD. Licensing options range from free access for academic purposes to enterprise-level solutions leveraged by major pharmaceutical companies worldwide.
Read about our drug, COVID-19, and pesticide data subsets that can further help focus computational research efforts.
Watch this presentation on how effective use of structural data can underpin the design of medicines and experimental and computational chemists work together to solve real-world pharmaceutical challenges.
Visit the WHO’s WAAW webpage for more information on this year’s campaign, which runs from 18–24 November. #WAAW