CSD Data Curation – the challenge of a million structures
In a recent blog - CSD Data Curation - The Human Touch - we’ve described the work that goes on when new structures are added to the Cambridge Structural Database (CSD). However, It’s important to realise that this isn’t the end of the story - as we get close to adding the one millionth structure to the CSD, it seems like an appropriate time to describe some of the processes we undertake at the CCDC to ensure that the data we make available to scientists continues to empower and inform their research long after it’s initial deposition.
Improvement projects to enhance entries
A few years ago I wrote a blog discussing the challenges we face when assessing unusual features of a crystal structure. Research is often reported precisely because the results are novel and unusual, however with a database of almost a million structures we need to try and represent data in a consistent way to give meaningful search results. At the time, in 2015, the CCDC had only recently released it’s Python API and we were exploring how best to employ the new capabilities it afforded to us to update and improve existing entries in the CSD.
The histogram below from Mercury’s Data Analysis Module hopefully helps to illustrate the challenge we face. This histogram shows the result of a ConQuest substructure search for the occurrence of the glycouril fragment (shown in the top left of the chart). The fragment was almost entirely absent from the CSD (other than a few sporadic reports in 1965 and the late 1970’s not included in the chart) until the publication in 1981 of a JACS paper titled ‘Cucurbituril’ by Freeman, Mock and Nhih (10.1021/ja00414a070). The Cucurbituril molecule from this publication (CSD refcode BEBDOB is shown in the centre of the chart) is a macrocycle formed from repeating glycouril units. Following its discovery it has since been studied extensively – now with over 100 new examples added yearly. Over time this new family of molecules has developed its own conventions – for example in how the compounds are named, and therefore historical entries in the CSD must be reviewed and updated to reflect these new conventions.
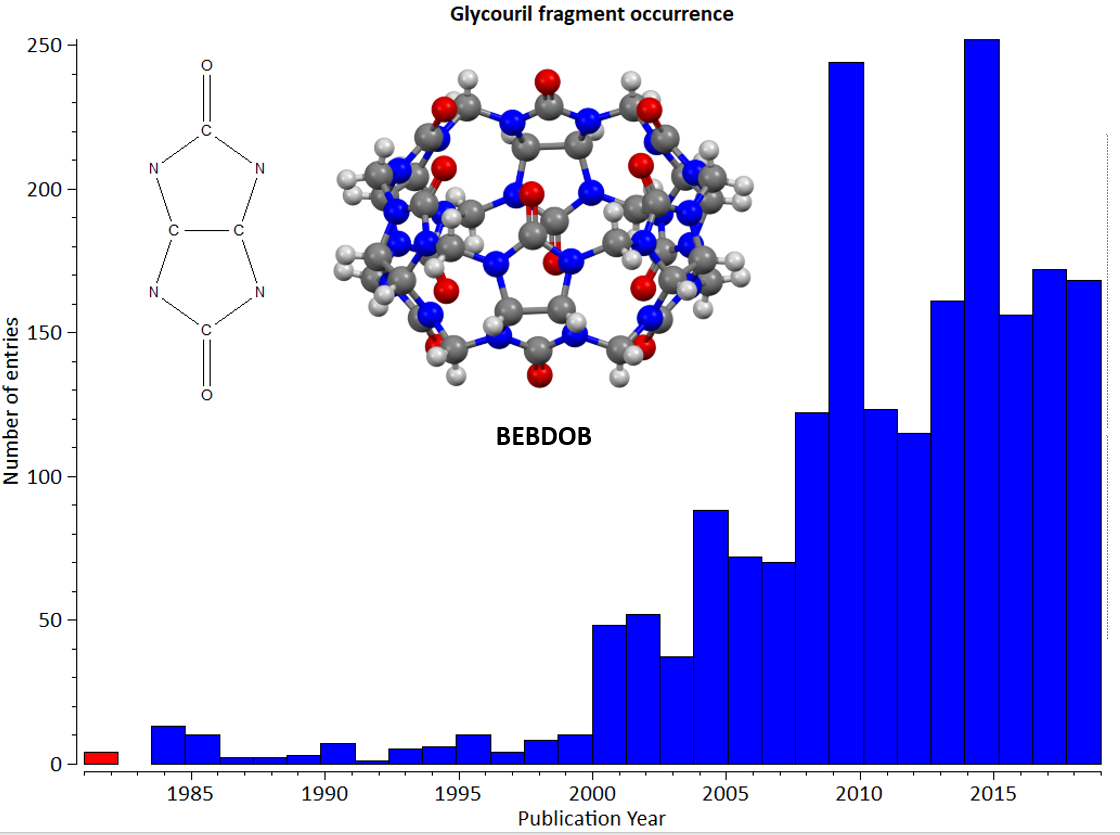
Histogram showing the increasing occurrence in the CSD of crystal structures including the glycouril fragment. The column including the first Cucurbituril structure (CSD refcode BEBDOB) is highlighted in red.
We now undertake a series of improvement projects on CSD entries every year, with some examples of the updates to the CSD reported with every CSD-System release. In the latest 2019 CSD, over 80,000 existing entries were updated in some way.
The focus of our improvement projects depends upon feedback we receive from our users; for example feedback on a particular entry to data_edits@ccdc.cam.ac.uk or from conversations at conferences or through research collaborations. We aim to prioritise our work on what will give the most benefit to users.
As an example, last year we used the CSD Python API to compare the molecular formula given in the CSD entry with the elements present in the atomic coordinates from the deposited CIF file. The aim of the project was to ensure any highly disordered molecules not modelled from the diffraction data were correctly reported in the CSD entry. Whilst this search enabled us to easily find such cases it also highlighted inventive approaches used by researchers to model their X-ray data. One such structure is CSD refcode ZIYPAY as shown below. The paper, from the Journal of Molecular Structure (10.1016/0022-2860(94)08405-7) investigated phase transitions in a diethylammonium hexabromo-tin structure. However, as the authors could not model the highly disordered diethylammonium cations, an isolated technetium atom (which is isoelectronic with the diethylammonium cation) was chosen to represent the cation instead.
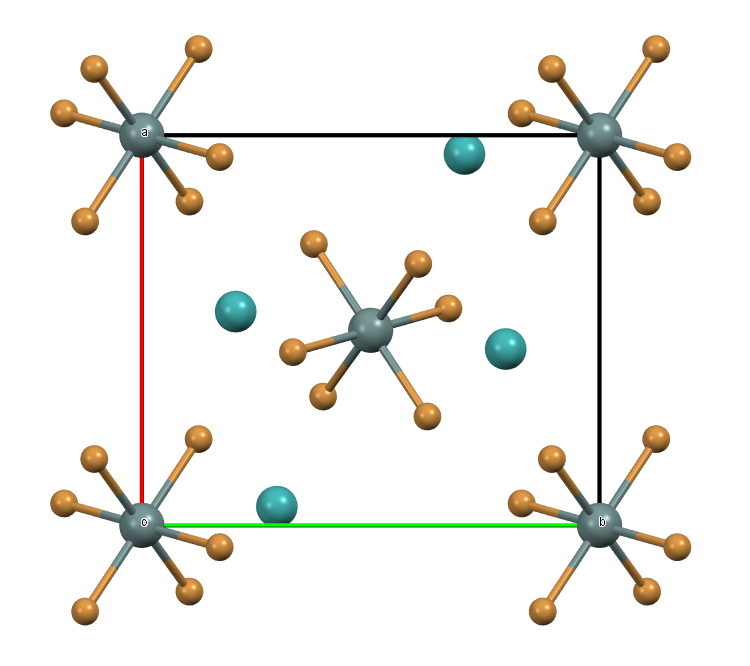 Unit cell of structure ZIYPAY viewed along the c-axis. The isolated Tc atoms (shown in turquoise) represent disordered cations in the structure.
Unit cell of structure ZIYPAY viewed along the c-axis. The isolated Tc atoms (shown in turquoise) represent disordered cations in the structure.
As well as improving how we represent and interpret existing entries our improvement projects also aim to enhance existing entries. These enhancements cover a wide variety of CSD entry features, from back-populating publication DOIs when journals make historical publications available online to a continuing multi-year project to add metal oxidation states to compound names, with over 100,000 additions so far. We’ve also looked at enhancing entries for well-known drug molecules with common names and melting points. All these enhancements are designed to increase the discoverability and usability of the data within the CSD – please do let us know what would be most impactful for your research.
This year we hope our improvement projects will have greater impact than ever, as we’ve recently welcomed a Post-Doctoral Research Scientist to work with us, with a specific focus on investigating the integrity of structural data in the CSD. We’ll cover this in more detail in an upcoming blog.
Looking further ahead we would like to use the experience gained from our own improvement projects to develop tools to help depositors when data is first deposited with us. Our online web deposit service already guides depositors through a simple multi-step process where they are given a chance to enhance and review their data; as we look forward to the next million CSD entries we’d like to improve this even further.