Bringing Order to Disorder: Expanding Structural Detail in the Cambridge Structural Database
Disorder is a common and important feature in crystal structures. It occurs when atoms or molecules do not occupy a single, well-defined position within the crystal lattice, but instead adopt multiple positions or orientations. This may arise from thermal motion, partial occupancy, or structural ambiguity and reflects the inherent complexity and flexibility of chemical systems.
We’ve been working hard to improve how disorder is represented in the Cambridge Structural Database (CSD). In this blog post, we’ll explore the improvements we’ve made, the challenges we’ve faced, and why accurately capturing disorder is essential for structural chemistry and data-driven research.
Disorder Handling Improvements
Over the past few years, we’ve enhanced how disorder is processed, stored, and visualized across the CSD. Our goal has been to ensure that all disorder models are faithfully captured, viewable, and searchable, rather than simplified or partially hidden.
As part of this effort, we’ve made updates across the CSD Portfolio, including Mercury and the CSD Python API. You can learn more about some of these changes in our previous blog posts:
- Handling Disordered Structures in Mercury and the CSD Python API
- How to Visualize and Handle Disorder in Mercury and the CSD
How Disorder Was Previously Represented
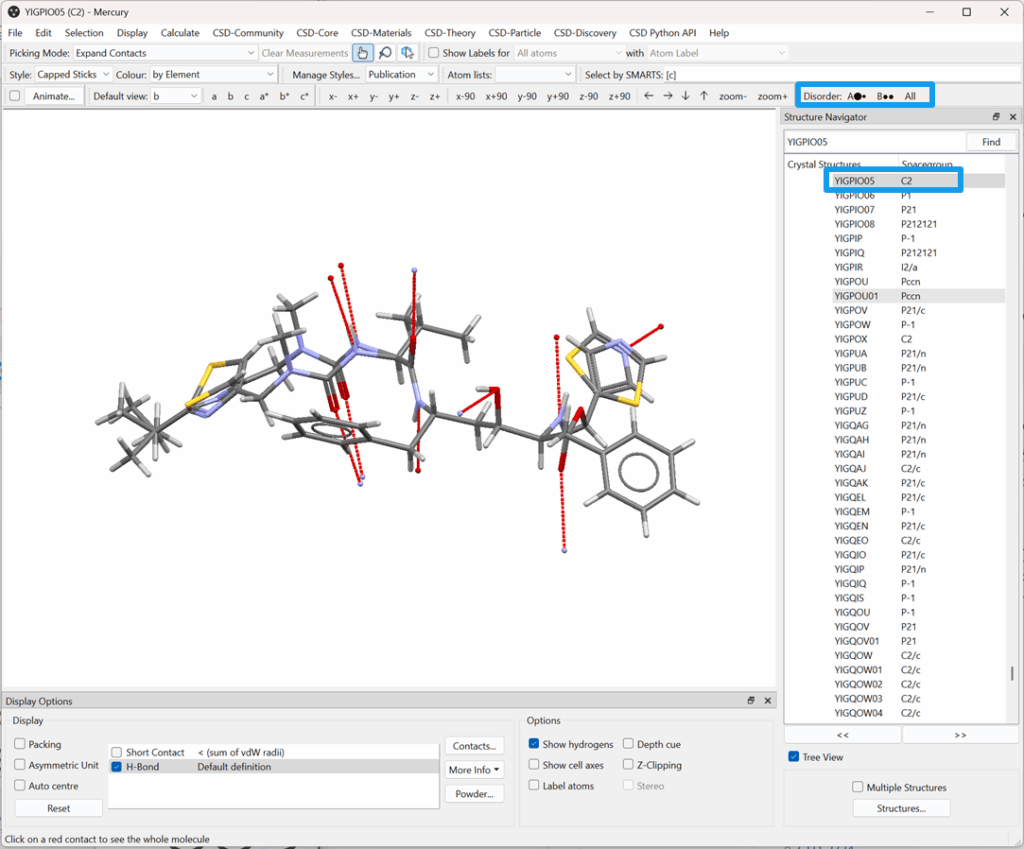
Historically, disorder in the CSD was simplified: only the major disorder component was fully represented in the 3D structure. Minor components were “suppressed” and visualized as pale green atoms which were visible but not interactive. It was not possible to view complete models for alternate disorder assemblies or toggle between them.
This approach allowed for cleaner visualization but often obscured valuable structural information. As a result, users couldn’t fully explore the true variability present in disordered structures.
Improved Disorder Models in the 2025.1 Data Release
With the 2025.1 data release, we’ve made a major step forward by replacing suppressed atoms with fully calculated disorder models in over 165,000 entries.
Now, when you open a disordered structure in Mercury:
- All disorder models are displayed by default.
- Use ‘Element or Disorder’ colouring to easily identify disordered atoms.
- The disorder toolbar shows assemblies (by letter) and disorder groups (by circles).
- Click the toolbar to toggle through the different disorder models.
To generate these new models, we extracted detailed disorder information from deposited CIFs and used automated methods to calculate all valid disorder representations. Importantly, we validated each new model against the previously accepted 3D structure, updating it only when we were confident that the major disorder model aligned with the original representation.
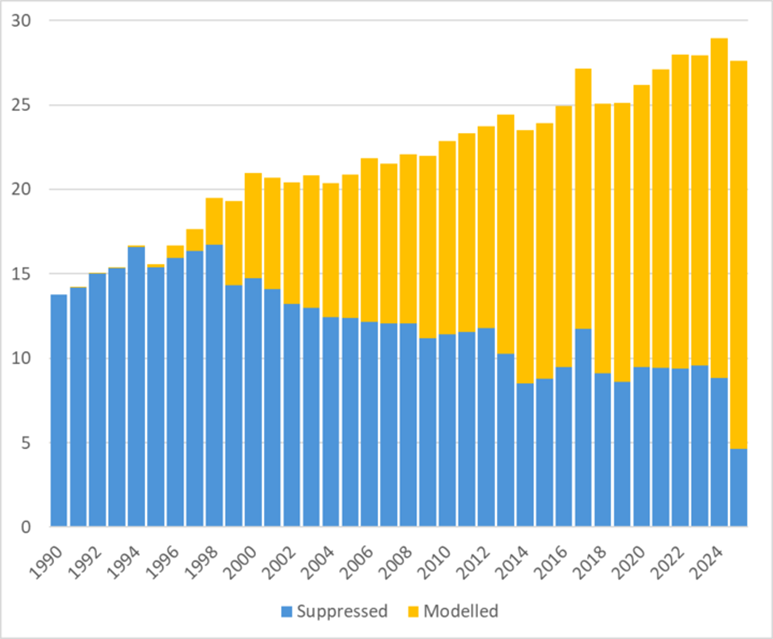
As shown in the graph below, validation success rates improve significantly with newer structures, with over 85% of entries published in 2024 successfully converted to validated disorder models.
Challenges in Calculating and Validating Disorder Models
Creating robust disorder models presented a number of technical challenges:
Inconsistencies in deposited CIFs – Disorder is expressed in varied and sometimes ambiguous ways, depending on the refinement software and crystallographer preferences.
Complex disorder cases – Including symmetry-related disorder, where disordered atoms are generated through crystallographic symmetry operations. These are particularly difficult to calculate and validate automatically.
Because of this, we excluded symmetry-related disorder from the first phase of back-population. However, we plan to tackle these more complex cases in future releases.
Benefits
The benefits of this work are already being felt:
- More accurate structural data – Users see what was truly observed, not a simplified model.
- Improved search and exploration – Structures with disorder can be filtered, compared, and analysed more effectively.
- Support for machine learning – Richer, more complete data supports AI and statistical analysis, where missing or simplified disorder can bias results.
- Better visualization and insight – Researchers can now fully explore alternate molecular configurations within a structure.
Ultimately, this leads to a richer, more representative database, enabling deeper chemical insights and more robust applications of the data.
Looking to the Future
Our work with disorder is ongoing. In the coming releases, we plan to:
- Expand coverage to more complex disorder cases, including symmetry-related disorder.
- Continue updating older entries where automated validation of disorder models previously failed.
- Extend support for disorder models across more tools in the CSD Portfolio.
- Enhance integration with search tools, allowing users to search by type and degree of disorder.
We’re committed to continuing improvements in this area and welcome your feedback. If you work with disordered structures, we’d love to hear what features or data would be most useful to you as we plan future development.
We hope you find these improvements useful in your research. Representing disorder more fully is a key step toward a more complete and chemically insightful CSD. If you have thoughts, requests, or feedback about disorder handling, please get in touch. Your input will help shape the development priorities for future releases.
Next Steps
To discuss further and/or request a demo with one of our scientists please contact us via this form or .
Check out the Cambridge Structural Database (CSD) for yourself and see how your research can benefit from the combined knowledge of over 1.3M small-molecule organic and metal-organic crystal structure data.
Proprietary data? Our team can curate your proprietary data into a Cambridge Structural Database (CSD)-like database, accessible through a simple browser-based interface. This is all done within your firewall to comply with your data security requirements.
More information on CSD software trusted by academic and industrial institutions around the world.
See case studies of the CSD in action, driving forward the boundaries of scientific research.