Fragment Hotspots
Fragment Hotspot App:
- Given a whole protein and no prior knowledge, the Fragment Hotspots application identifies the location and quality of binding sites in minutes.
- Not only do high scoring regions coincide with ligand binding site, but the very highest scoring regions specifically highlight fragment binding sites.
Whitepaper: Understanding Drug Selectivity and Pocket Druggability with Ensemble Hotspot Maps
Identifying the interactions that determine fragment binding:
Locating a ligand-binding site is an important first step in structure-guided drug discovery, but current methods do little to suggest which interactions within a pocket are the most important for binding. Together with one of our industrial collaborators we have developed a method that samples atomic hotspots with simple molecular probes to produce fragment hotspot maps. These maps specifically highlight fragment-binding sites and their corresponding pharmacophores. The fragment hotspot map calculation is validated using experimental binding positions of 21 fragments and subsequent lead molecules. The ligands are found in high scoring areas of the fragment hotspot maps, with fragment atoms having a median percentage rank of 97%. Protein kinase B and pantothenate synthetase are examined in detail. In each case, the fragment hotspot maps are able to rationalise a Free–Wilson analysis of SAR data from a fragment-based drug design project. For more information see J. Med. Chem., 2016, 59 (9), pp 4314–4325 DOI: 10.1021/acs.jmedchem.5b01980.
Applications of Fragment Hotspot maps:
- Identify ligandable binding sites,
- Predict which interactions fragments are most likely to make,
- Identify docking constraints when working with a novel pocket or target,
- Refine your pharmacophore to target the most valuable interactions during a virtual screen,
- Visualise and guide fragment growth or lead optimisation.

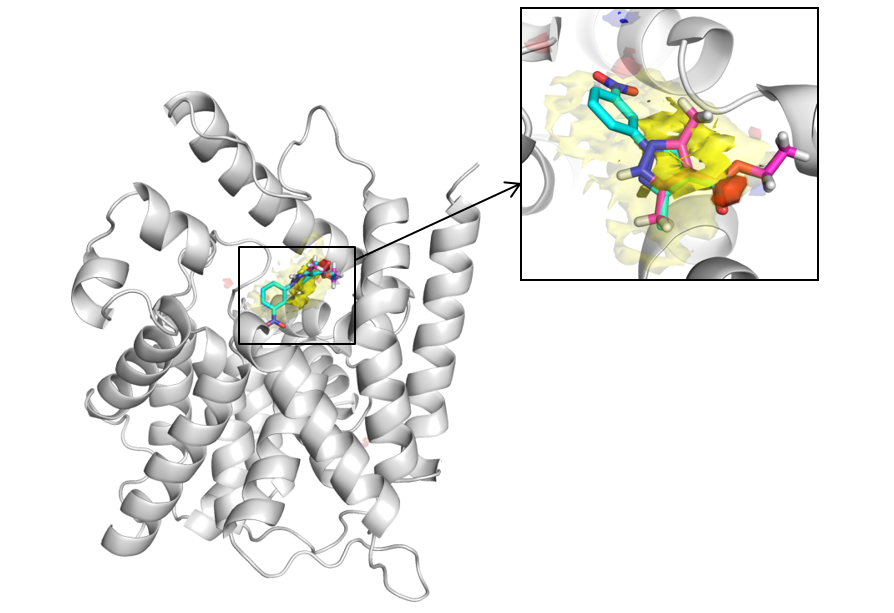
Apo PDE4 with apolar (yellow), donor (blue) and acceptor (red) maps contoured at 17 (solid) and 14 (translucent). The binding position of a fragment (magenta) and its subsequent lead (cyan) molecules have been included for reference. From a global search of the protein, the Fragment Hotspot Maps are able to highlight the interactions made by the fragment, without false positives elsewhere on the protein.
Reprinted with permission from J. Med. Chem., 2016, 59 (9), pp 4314–4325 DOI: 10.1021/acs.jmedchem.5b01980. Copyright 2016 American Chemical Society.