Will electron diffraction revolutionize drug development?
Q&A with Dr Jessica Bruhn, Scientific Group Leader - MicroED at NanoImaging Services about a new advancement in drug development
Microcrystal electron diffraction (MicroED or 3D ED) uses a beam of electrons rather than X-rays to obtain structures. In a recent paper, a team of researchers led by NanoImaging Services presented a new advancement in drug development using electron diffraction for when X-ray diffraction (XRD) may not be an option. In this blog, we talk with author Dr Jessica Bruhn, Scientific Group Leader - MicroED at NanoImaging Services about the team’s findings.
Dr Bruhn and team recently published the paper, Small Molecule Microcrystal Electron Diffraction for the Pharmaceutical Industry – Lessons Learned from Examining Over Fifty Samples, in the journal Frontiers in Molecular Biosciences.
In 10 years, what do you think the ratio between 3D ED structures and X-ray structures will be?
I think structures determined by X-rays will still far outnumber those determined by electron diffraction, even in another ten years. Single crystal X-ray diffraction is faster, cheaper and easier to access compared to electron diffraction today, and probably still will be in ten years. I do hope/expect to see electron diffraction determining more and more structures inaccessible to X-rays, such as those of transient polymorphs, helping to expand the breadth of crystal structures that can be determined. It will also be interesting to see if 3D ED replaces structure determination from X-ray powder diffraction. Anything that diffracts in a powder diffractometer should be suitable for 3D ED, which arguably provides higher quality data that is easier to process. I don’t expect to see powder diffraction disappear, especially given how valuable this technique is for analytical purposes, but I do wonder if we will see fewer structures produced from powder as 3D ED grows.
Could 3D ED one day overtake single-crystal X-ray diffraction experiments?
It is certainly possible. Growing large crystals is a huge bottleneck for those interested in determining crystal structures. ED can work with crystals of almost any size as it is generally fairly straightforward to break large crystals into a size suitable for ED. In many cases, crystallization experiments can be completely eliminated as many compounds spontaneously form crystals during purification. I could see advances in the design of sample holders/goniometers that could really push this technique to be the method of choice.
Will 3D ED adoption rates differ between academia and industry?
I think that this will depend on how long it takes for more microscopy facilities to start offering 3D ED services. Most academic institutions and many large pharmaceutical companies have access to microscopes capable of collecting ED data, but very few institutions and companies have scientists who have been trained to collect and process such data. The demand for 3D ED structures is quite large, but the capacity is really lacking at the moment. In the short term, I see 3D ED growing faster in industry where they have the financial resources to access service providers, such as NanoImaging Services. For academia, I think it just depends on how quickly each individual institution can get an ED center up and running, and how they are able to subsidize the high cost of operating these microscopes.
What are the current limitations and challenges with 3D ED and how quickly is the field evolving to overcome these?
Just about every time I talk to a chemistry group about adding 3D ED to their workflow, absolute configuration gets brought up. Absolute configuration refers to our ability to determine the handedness of chiral compounds. Most small molecule drugs are chiral, with generally only one form being active. So, understanding which enantiomer is biologically active is of critical importance to those in the pharmaceutical space. Single crystal X-ray diffraction can regularly and reliably provide this information. Things are a little more challenging in electron diffraction, but I am closely watching groups such as Lukas Palatinus’ at the Institute of Physics of the Czech Academy of Sciences as they come up with clever solutions to this problem. I am hopeful that absolute configuration can soon be easily determined from 3D ED data.
I am also really looking forward to seeing more advances on the data processing side of things. 3D ED data poses some unique challenges compared to typical single-crystal X-ray data, such as the need to combine data from multiple crystals, dealing with radiation damage, indexing ambiguities and the challenges of phasing lower resolution data (>1.1 Å). Many of these challenges are shared in the protein crystallography field, and I am hopeful that we can look to the many decades of innovation in the protein field for inspiration as we work to build better tools for 3D ED. The real challenge here will be demonstrating that there are enough structures being determined by 3D ED that it is worth the time and effort of software developers to build 3D ED specific tools. I am hopeful that our paper and others in the 3D ED special collection at Frontiers in Molecular Biosciences are helping to encourage software developers to do just this.
Read more
We hope you have enjoyed learning from Jessica Bruhn, an expert on MicroED, about how her field may evolve and impact the Cambridge Structural Database (CSD) in the coming years.
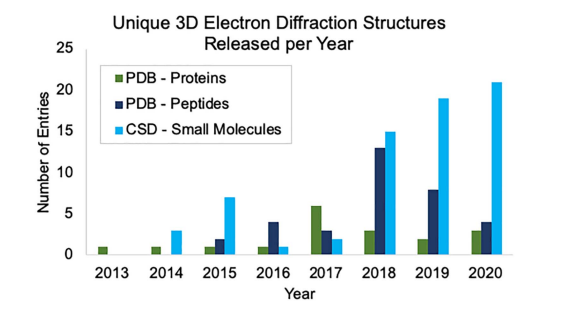
To learn more about MicroED, read the full paper here, which includes metrics on the recent growth of 3D ED in databases like the CSD and the Protein Data Bank (PDB).
Our press collaboration about the paper is also available here.
For more information on how to access and deposit electron diffraction structures in the CSD, read this blog by CCDC Data Integrity Research Scientist, Natalie Johnson on Electron Diffraction Data in the CSD.