Visualizing Crystal Interactions
You can now quickly identify and visualize the interactions that hold crystal structures together using new functionality in Mercury and the CSD Python API.
Interaction networks use intermolecular interactions to construct Periodic Bond Chains (PBCs) according to the theory first introduced by Hartman and Perdok. PBCs are chains of repeating strong interactions that can be used to explain the relationship between crystal structure and crystal growth or to aid the interpretation of other properties of a crystal structure.
Interaction networks in Mercury enable the visualization of individual bond chains, or to build a network of interactions by selecting multiple PBCs. The rules for the PBC search can be adjusted, and PBCs can be grouped by direction and/or symmetry.
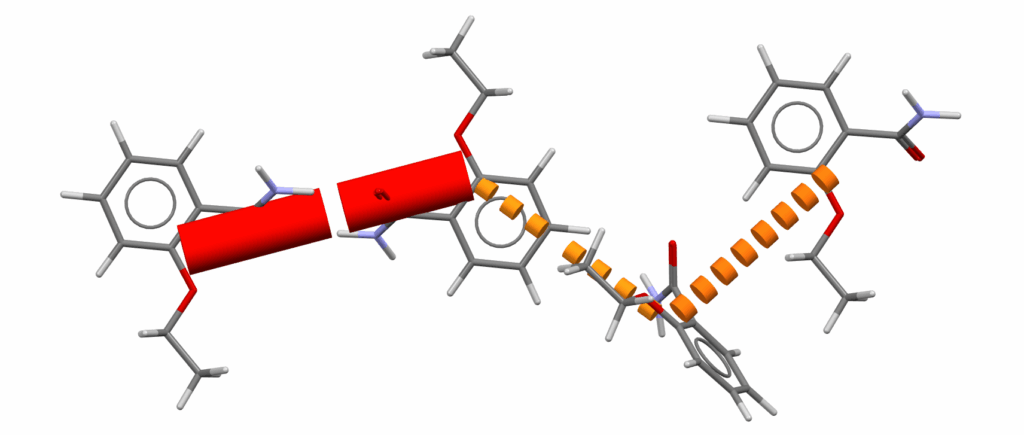
The Mercury dialog offers a powerful range of visualization options, enabling compelling graphics to be created. Users can choose how to represent strong and weak bonds using colour, dashes, line thickness, or a combination of all these elements.
In combination with Mercury’s style menu, further refinements can be made — the ‘Work’ style provides simple and easy-to-interpret lines, the ‘Publication’ style displays elegant cylindrical links. Labels can be shown to add information or removed for clarity, and molecules can be hidden to abstract the structure to the intermolecular interactions.
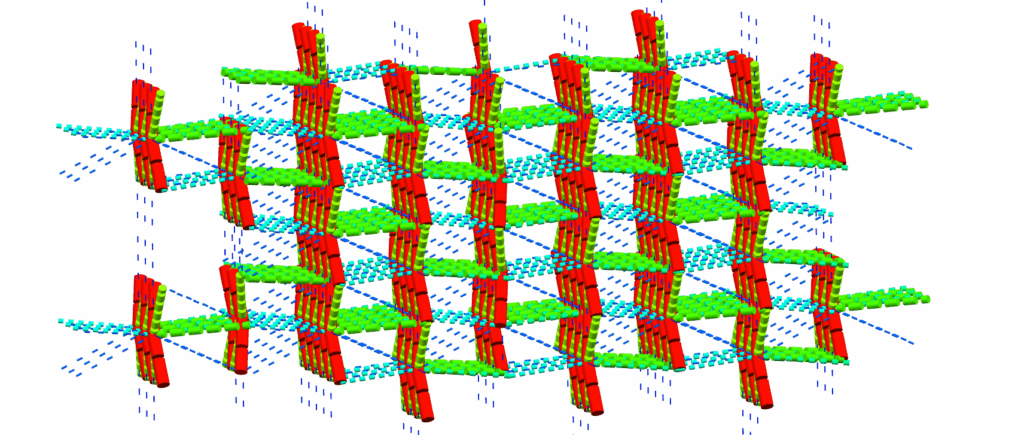
The dialog identifies the plane(s) that combinations of PBCs form, and provides easy-to-use links to filter the PBC list to just one plane of interest. The selected layer can then be easily sliced, or a surface analysis can be performed to visualize the properties of the potential face containing these PBCs (CSD-Particle licence required for the latter).
Three force fields can be used in the calculations — CLP [1, 2, 3], UNI [4, 5] and Cambridge Structural Database – Optimized Potential for Crystal Structures 2016 (CSD-OPCS16), also used in the Landscape Generator [6].
The interaction networks dialog checks that the structure being assessed is calculation-ready (e.g. no missing hydrogen atoms) and that the force field selected is parameterized for the chemistry in the structure.
The interaction networks functionality is available in Mercury and the CSD Python API to users with a CSD-Materials or CSD-Enterprise licence.

The interaction network functionality was developed as part of the DigiCCAMMS program, in partnership with the University of Durham. This project will integrate the ADDICT prototype into the CSD-Particle suite using Innovate UK funding as part of the Sustainable Medicines Manufacturing Innovation: Collaborative R&D competition.
Next Steps
Contact us here to discuss further and/or request a demo with one of our scientists.