Three things solid form informatics can tell you about your drug candidate
Solid form informatics allows researchers to assess exciting therapeutic candidates. By analyzing intra- and intermolecular interactions, geometries, and crystal structure packing, researchers are better able to understand the challenges associated with a potential new active pharmaceutical ingredient (API)—like polymorphism and manufacturing risks—in order to ensure the best candidates progress to the next stage of the development process. In this blog, we’ll look at three key insights that the CCDC’s solid form informatics approach can tell you about your drug candidate.
How can solid form informatics highlight key properties like stability, dissolution, and bioavailability?
Solid form informatics can distil the information contained within crystal structure data into insights that rationally guide drug design and development. CCDC’s CSD-Materials software package leverages the Cambridge Structural Database (CSD), along with in-house structural databases, to help you optimize the solid form properties of a potential new API or engineer new compositions. Specifically, CSD-Materials can help you:
- Compare an experimental or predicted structure’s important geometries—including bond lengths, angles, ring geometries, and torsions—to those found in structural databases.
- Identify the likelihood and strength of all hydrogen-bonding networks and understand any unusual geometries of an observed hydrogen-bonding network, as it compares to other relevant known structures in the CSD.
- Determine a molecule’s preferred interactions and quickly evaluate the intermolecular packing of the crystal structure via an intuitive 3D visualization.
Geometry check: identify preferred molecular geometries with CCDC’s Mogul component
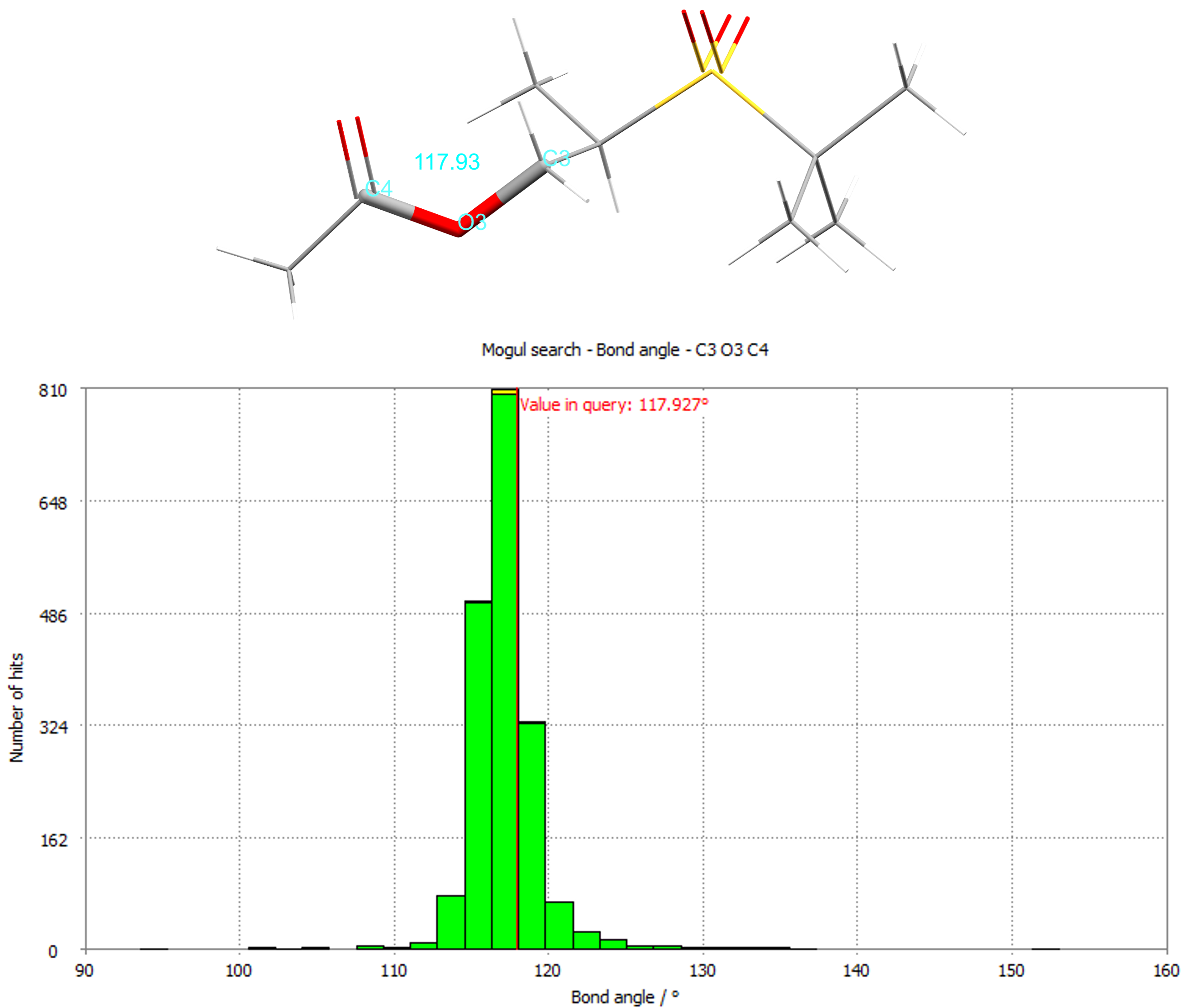
The bond angle histogram for the C3-O4-C4 angle for CSD refcode NOBWIK.
By identifying unusual features, like uncommon torsions or bond angles, you can de-risk a potential new structure. The CSD-Materials component Mogul derives the quality of intramolecular geometries and can display distributions of important geometries based on the bond lengths, angles, ring geometries, and torsions of the expertly curated structures in the CSD.
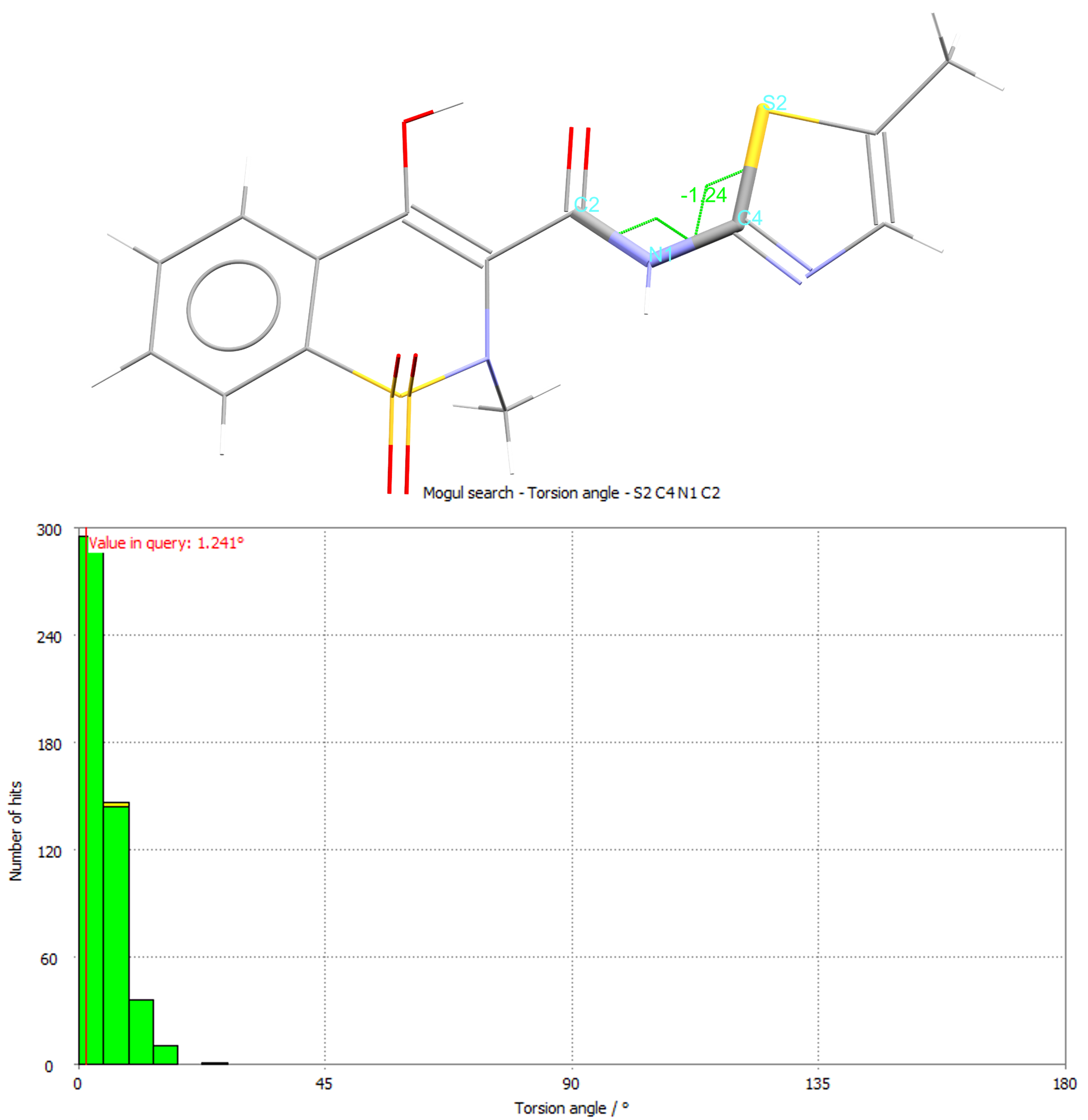
The histogram from the torsion for the angle highlighted in green for CSD refcode SEDZOQ.
It’s a great way to assess the geometry of experimental or predicted structures and predict the likely conformations of chemistry before a structure has even been determined! You can also drill down into each report to see the individual structures driving the results, which can include up to 10,000 experimentally derived and peer-reviewed structures housed in the CSD.
- Read how a researcher at Astex Pharmaceuticals used Mogul to analyze the geometry of drug-like ligand models for high-resolution crystallographic protein-ligand complexes.
- View our free, on-demand tutorial on how to run a Mogul geometry check.
Intermolecular analysis: understand stability through Hydrogen Bond Propensity and Hydrogen Bond Statistics
CSD-Materials has multiple components to help you understand how potential hydrogen-bonding networks may affect stability.
Compare the relative likelihood of potential hydrogen-bonding networks with Hydrogen Bond Propensity (HBP)
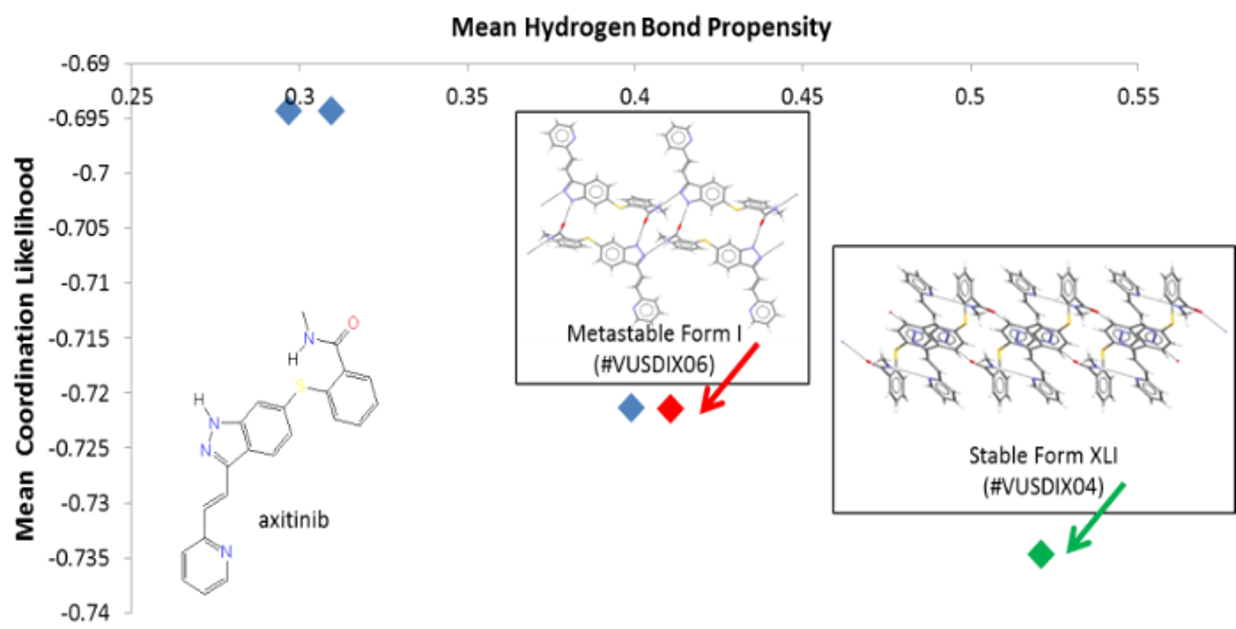
An example of how to assess the risk of polymorphs using an HBP analysis. This graph is for the putative hydrogen-bonding networks for axitinib, a chemotherapy drug. Experimentally validated forms of axitinib that are found in the CSD, refcodes VUSDIX06 and VUSDIX04, are highlighted in red and green, respectively.
HBP can help predict likely hydrogen bonds for a given molecule, assess crystal forms by identifying sub-optimal hydrogen bonding, calculate hydrogen bond propensities for individual donor and acceptor group pairs, and perform a comprehensive analysis of hydrogen bonding on a set of structures.
- Learn more about HBP and its important role in derisking metastable solid forms.
- Read how a research team used HBP to characterize two polymorphs of an anti-inflammatory drug and predict the existence of additional forms.
Understanding the geometry of hydrogen-bonding networks with Hydrogen Bond Statistics
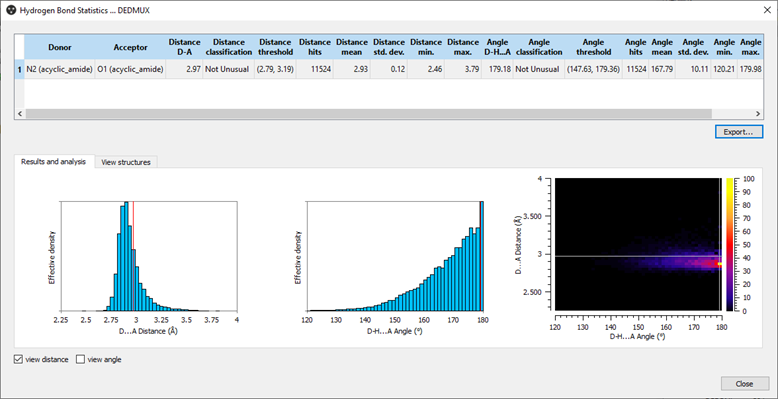
Hydrogen Bond Statistics for CSD refcode: DEDMUX.
Hydrogen Bond Statistics allows you to analyze how typical the geometry of an observed hydrogen bond in a crystal structure is. It compares the interaction geometries from your predicted or experimental structure to the over 1.1 million crystal structures in the CSD so that you identify which hydrogen bonds may have a stabilizing effect. The easy-to-read histograms and charts place your structure relative to others, and you’re able to view the individual structures driving the graphs as well.
What if my solid form doesn’t have hydrogen bonding?
Hydrogen bonding can play a key role in identifying the risk of metastable polymorphs. But what if a system doesn’t have hydrogen bonding or the hydrogen-bonding networks are the same in two different solid forms? The CSD-Materials suite goes beyond hydrogen bonding to look at other factors, like aromatic interactions.
Packing analysis: determine a molecule’s preferred interactions with Full Interaction Maps (FIMs)
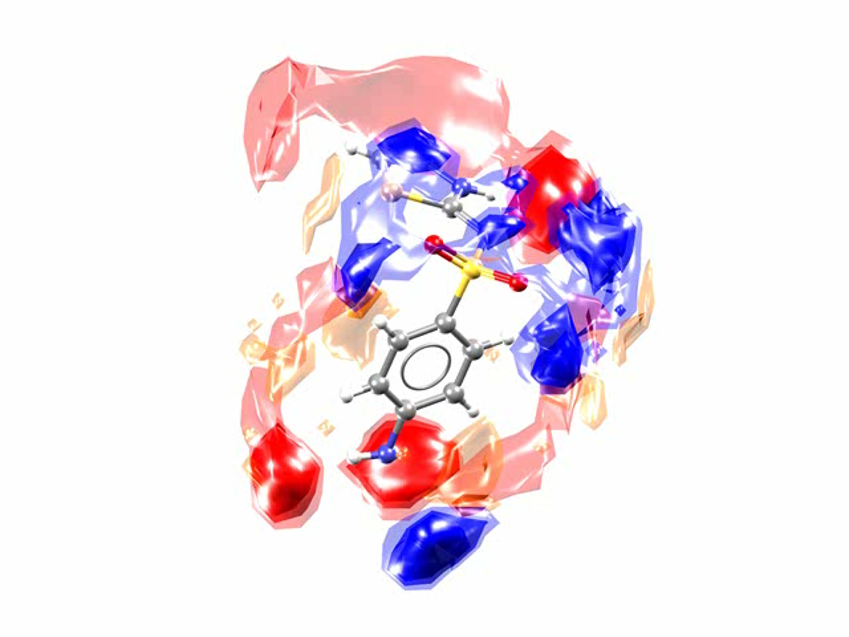 This is the Full Interaction Map for the SUTHAZ family of structures. The map compares the observed intermolecular interactions with preferred geometries for that type of interaction. The red regions of the map denote areas where there is a high probability of a hydrogen-bond acceptor, the blue regions denote likely hydrogen-bond donors, and the yellow regions indicate potential interacting aromatic groups.
This is the Full Interaction Map for the SUTHAZ family of structures. The map compares the observed intermolecular interactions with preferred geometries for that type of interaction. The red regions of the map denote areas where there is a high probability of a hydrogen-bond acceptor, the blue regions denote likely hydrogen-bond donors, and the yellow regions indicate potential interacting aromatic groups.
FIMs provide your molecule’s interaction preferences in the context of the observed crystal structure in an intuitive, 3D visualization. From this, you can assess the different interactions that occur in observed polymorphs, identify regions where a co-former interaction may result, explore the likely interactions of a molecule prior to crystallization, and learn where a solvent or an inhibitor may interact.
- Read more about the science behind FIMs using three pharmaceutical compounds: sulfathiazole, anastrozole and cipamfylline.
- Watch our free, on-demand tutorial on how you can analyze intermolecular interactions for small molecules with FIMs.
Learn more
Solid form informatics provides a knowledge-based approach to improve the effectiveness and quality of early-phase drug discovery research, as well as a rational approach to risk assessment. Want to learn more about how it can help your solid formulations for drug research?
Download our free whitepaper: “Solid Form Informatics for Pharmaceuticals and Fine Chemicals“
.wmv)