New Structures and New Data Fields. The Latest CSD Data Update
The Cambridge Crystallographic Database (CSD) has been fully updated, adding the latest structures as well as improvements to older structures. An additional 17,681 structures have been added to the database, bringing the total to 1,374,731 unique structures (1,413,222 entries).
In addition to the new structures, the database has been further enhanced with important new fields, searchable in the CSD Python API and visible in Mercury. These fields include Wavelength, Resolution, Flack Parameter and Data Availability. See the blog ‘Wavelength, Resolution, Flack Parameter and Data Availability Indicators – New CSD Data Fields.’
The CSD 6.01 comprises 645,235 (46%) organic-only entries and 767,987 (54%) metal-organic entries from various sources including publications, patents and thesis data. The variety of chemistry contained within the CSD is just as broad, covering areas such as catalysis and pharmaceuticals to storage materials and semiconductors. Below we highlight some of the latest structures added to the CSD.
This release includes a study on the solid-state forms of elafibranor reported by Pellegrini et al. in Crystal Growth and Design. Elafibranor is a dual PPARα/δ-agonist drug used in the treatment of primary biliary cholangitis. This work elucidates the structures of two anhydrate polymorphs, a hydrate and several solvates, investigating the relationship between the different forms.
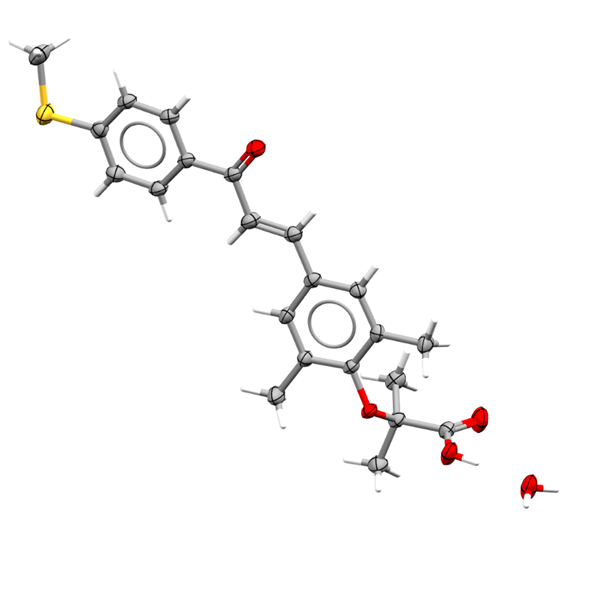
Also included is a study of high energy density molecules tailored towards molecular solar thermal energy storage (Biebl et al., Chemical Science; High energy density dihydroazaborinine dyads and triad for molecular solar thermal energy storage). This study explored the synthesis and reversible photoisomerization of azaborinines into 2-aza-3-borabicyclo[2.2.0]hex-5-enes with high energy densities being reported and improved UV absorption compared to previous iterations.
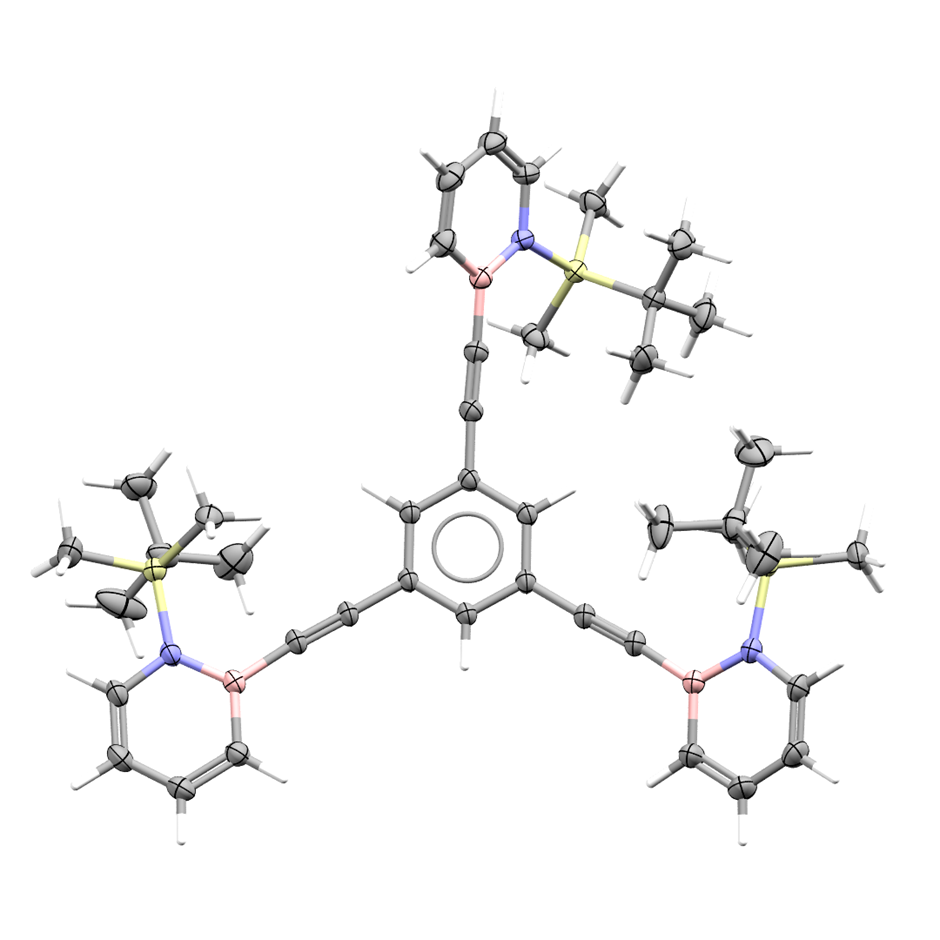
The variable oxidation state of germanium has been exploited by Zhou et al. in catalysis (Journal of the American Chemical Society; Germanium-Mediated Catalysis via Ge(II)/Ge(III)/Ge(IV) or Ge(II)/Ge(IV) Redox Cycling ). This study prepared a carbodiphosphoranyl germanium(ii) complex and applied it to hydrogenation and dearomatization reactions by virtue of different Ge redox cycles.
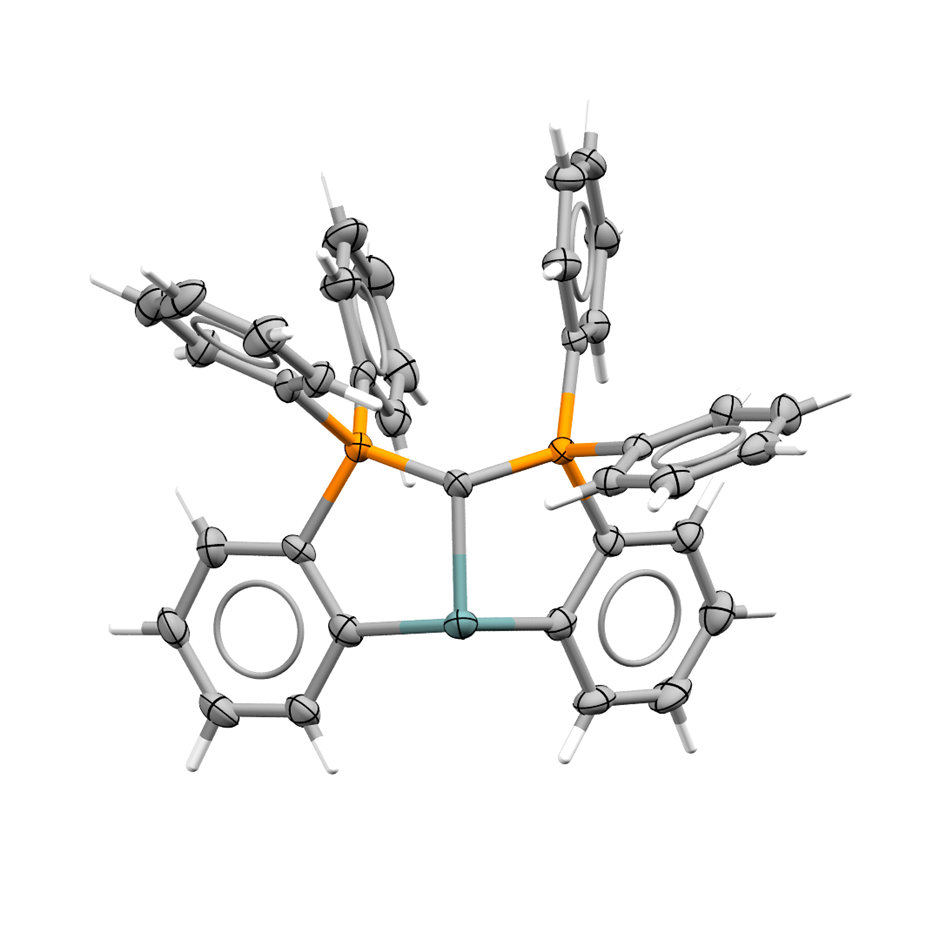
The chemistry of transuranium complexes has been investigated by Beck et al. and published in Nature Communications (https://doi.org/10.1038/s41467-025-63129-3). This study compared the bonding of maleonitrile-1,2–dithiolate with several lanthanide and actinide metal centres in terms of f-orbital participation and bond covalency. This publication adds to the rare examples of curium and californium in the CSD, bringing the total of each to 16 and 15 entries respectively.
This update also features new metal organic frameworks (MOFs). One way to filter the CSD to focus on MOFs is to take advantage of CSD subsets. In this way, you can restrict searches to specific MOF dimensionalities: 1D MOFs (37,938 entries), 2D MOFs (28,452 entries), 3D MOFs (33,656 entries) as well as non-disordered MOFs (100,556 entries).
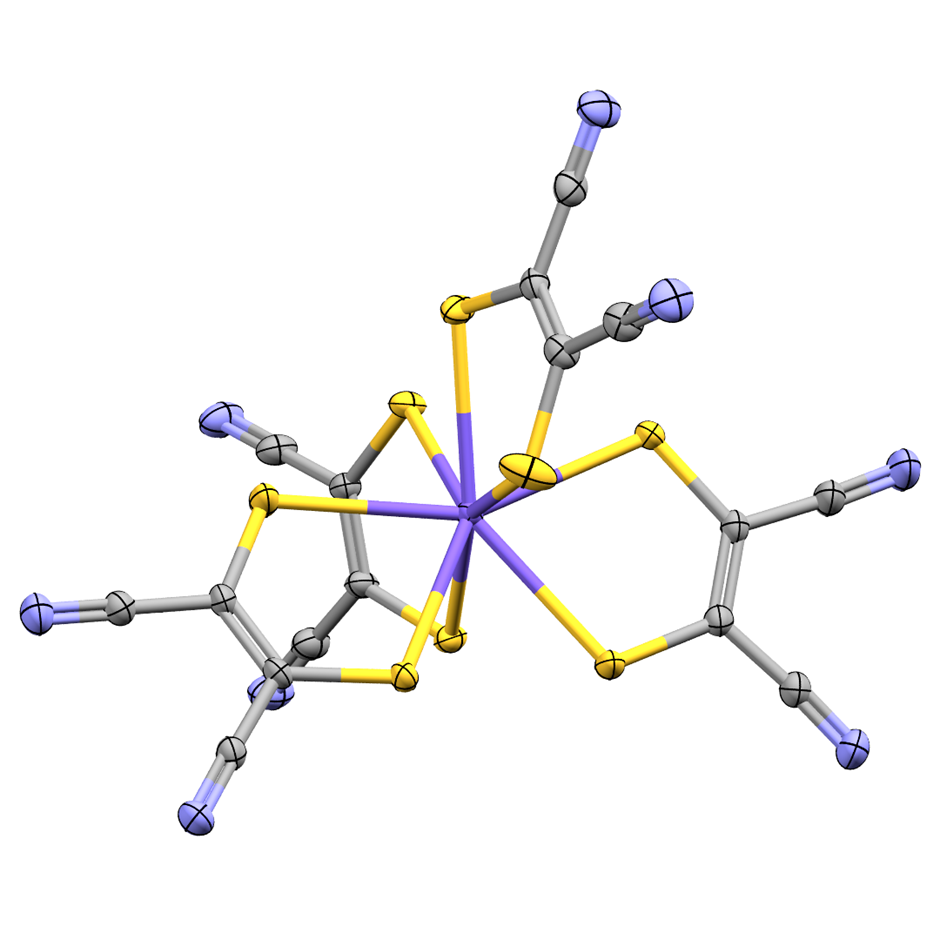
An example of a new addition to these subsets, appearing in the 3D and non-disordered MOF subsets has been reported by Zhang et al. in Angewandte Chemie (https://doi.org/10.1002/anie.202507349). One of a series of isostructural MOFs with an unh topology, JNU-300-Co was shown to activate 3O2 to 1O2 without the need for light irradiation at room temperature and have antibacterial activity related to this ability.
The CCDC also offers a pathway to share crystal data without a publication. These entries are published as CSD Communications directly through the database and there are over 69,000 of these entries in total in CSD 6.01. A new addition for this release is 41 entries from Judith Howard, who provided pdf files of her unpublished crystallographic data. These entries were converted into cif files by this year’s CCDC Summer Students.
In terms of data improvements added in this version of the CSD, further entries have been enriched with oxidation states, bioactivity information and radiation source labeling and this work was also carried out by this year’s Summer students. Following on from our release of disorder models to the CSD, we have also dedicated time to adding further disorder assemblies to entries. Over the next year, we intend to make improvements to linking between structures within the CSD and increase the number of entries with disorder assemblies.
New Data Fields — Wavelength, Resolution, Flack Parameter and Data Availability
It is not just new experimentally determined structures and improvements you have to look forward to in CSD 6.01. We are also delighted to bring you new data fields within entries. To read more about the new fields we’ve added, see the blog ‘Wavelength, Resolution, Flack Parameter and Data Availability Indicators – New CSD Data Fields.’
Next Steps
Do you have an improvement, suggestion or feature you would like to see for entries in the CSD? Get in touch and we’ll take them into consideration for our next data roadmap.