CSD-Discovery in Action: Identifying Aldose Reductase Inhibitors from Green Fluorescent Protein to Treat Diabetes Complications
Here we highlight a paper using CSD-Discovery to identify drug models for the treatment of complications due to diabetes and to define molecular features that can guide future drug design. This is part of our series highlighting examples of the Cambridge Crystallographic Data Centre (CCDC) tools in action by scientists around the world.
Summary
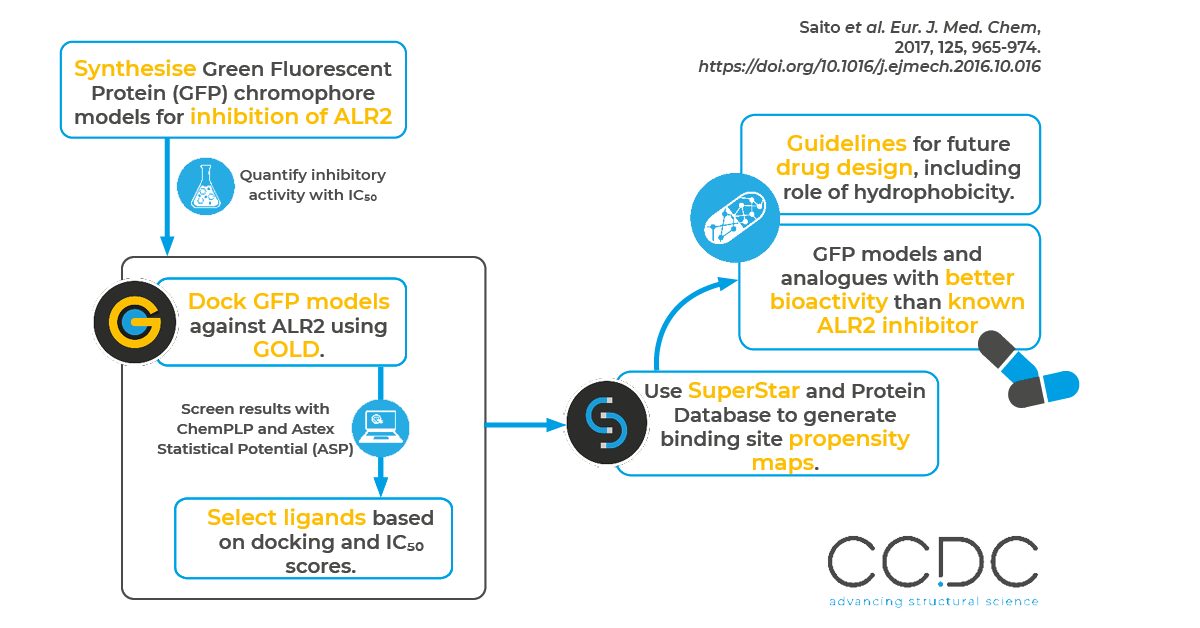
In this work, authors from Toho University and Kowa Company, Ltd used CSD-Discovery to evaluate the aldose-reductase-inhibiting properties of synthesized Green Fluorescent Protein (GFP) model compounds and their analogues.
Aldose reductase inhibitors are used to treat complications associated with diabetes. The authors confirmed the bioactivity of GFP chromophore models by measuring their IC50 values. One of the GFP chromophore models and its analogues exhibited very strong bioactivity, with efficacies higher than that of Sorbinil – a known, highly potent aldose reductase inhibitor. Along with their bioactivity study, the authors performed docking simulations in GOLD and then confirmed docking poses using SuperStar to define the interaction mode of the newly synthesized inhibitors toward the target protein and to identify the molecular features required to gain high inhibitory activity.
Why
Aldose reductase (ALR2) is an NADPH-dependent enzyme that catalyzes D-glucose reduction to D-sorbitol using NADPH as a reductant in the polyol pathway. Most glucose is metabolized via the tricarboxylic acid (TCA) cycle in the glycolytic system. But in patients with hyperglycemia, glucose actively fluxes into the glycolytic system and into the polyol pathway – where ALR2 is activated to consume the flooded glucose. As a result, high levels of D-sorbitol are formed intracellularly. The oxidation of D-sorbitol to D-fructose is very slow, which causes the intracellular concentration of D-sorbitol to increase and a large amount of NADPH is consumed. This results in an osmotic pressure imbalance that can cause diabetes complications, like peripheral neuropathy, cataracts and damage to the kidneys and small blood vessels. The inhibition of ALR2 activity may alleviate or prevent such complications.
Various types of ALR2 inhibitors (ARIs) have been reported and tested in clinical trials, but many show low activity or cause undesirable side effects, and such medications are fairly rare. For example, only Epalrestat was available as a therapeutic for diabetic peripheral neuropathy in Japan as of the authors’ work.
How
The authors chose the GFP chromophore model structure because it contains an N-carboxymethyl (NCH2CO2H) fragment similar to other ARIs and has a similar molecular skeleton and electronic structure to Epalrestat.
They used the GFP chromophore model to synthesize imidazolones and then measured the in vitro ALR2 inhibitory activity. They completed docking experiments with GOLD on 13 compounds into seven different protein structures obtained from the Protein Data Bank (PDB). This helped them determine the best pose for each experimental ligand. Then, using SuperStar, the authors generated interaction maps within the protein binding sites based on crystal structures in the PDB. For example, they were able to visualise the knowledge-based protein-ligand interaction area within the active site of one of the proteins (PDB ref code: 4IGS) to verify the role of hydrophobicity in the experimental ALR2 inhibitory activity in six of the 13 experimental ligands and analogues.
Read More
Read the full paper: “Green fluorescent protein chromophore derivatives as a new class of aldose reductase inhibitors” Eur. J. Med. Chem, 2017, 125, 965-974.
Learn more about CSD-Discovery.
Explore more examples of CCDC tools in action.