CSD Ligand Overlay in Action: Ligand-based Drug Design of Competitive Inhibitors Against DAHPS and EPSPS
Here we highlight a paper using the CSD Ligand Overlay tool in ligand-based drug design for the development of antibiotics and antifungals. This is part of our series highlighting examples of the Cambridge Crystallographic Data Centre (CCDC) tools in action by scientists around the world.
Summary
In this work, authors from the Universidade Federal do Pará used the CSD Ligand Overlay to develop pharmacophores that may inform new bioinspired competitive inhibitors with pharmaceuticals applications. Phosphoenolpyruvate (PEP) is a substrate for two catalysts involved in the shikimate pathway, which is essential for plant and fungi metabolism. The researchers used the Ligand Overlay tool to determine pharmacophoric groups of selected ligands that complex with PEP that could act as competitive inhibitors against two key PEP catalysts: 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHPS) and 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS).
They identified consistent binding modes of the selected ligands and identified pharmacophoric properties related to multi-target inhibitors for both enzymes. They found a ligand model that exhibited interesting binding energy with EPSPS, one showing an interesting inhibitor forming H-bond interactions with key-residues of PEP and a multi-target inhibitor for both DAHPS and EPSPS.
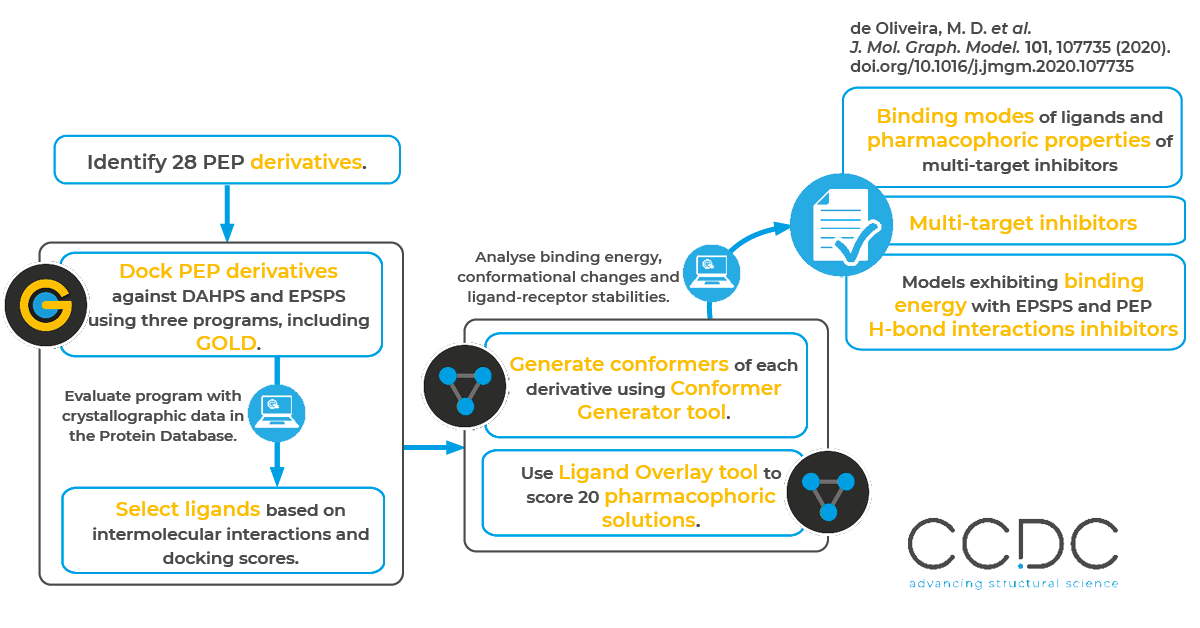
Why
The shikimate pathway is involved with the production of aromatic amino acids, such as phenylalanine, tyrosine and tryptophan – essential for plants, bacteria and fungi metabolisms. DAHPS and EPSPS catalyze important steps in the shikimate pathway using PEP as a substrate. New bioinspired competitive inhibitors against DAHPS and EPSPS could therefore support the development of therapeutics, like antibiotics or antifungals.
How
The researchers performed a literature survey of 28 PEP derivatives, and then analyzed the selectivity and affinity of the compounds against EPSPS and DAHPS using molecular docking, pharmacophore prediction, molecular dynamics (MD) simulations and binding free energy calculations. As part of the process, they used the CSD Ligand Overlay tool to determine pharmacophoric groups of the selected ligands that complex with both EPSPS and DAHPS. Specifically, they generated conformers for each PEP derivative using the Conformer Generator tool and then performed a pharmacophoric prediction using the Ligand Overlay application. The researchers optimized the overlay results based on volume, hydrogen bonding, hydrophobic coplanarity and internal energy scores.
Read More
Read the full paper: de Oliveira, M. D. et al. Targeting shikimate pathway: In silico analysis of phosphoenolpyruvate derivatives as inhibitors of EPSP synthase and DAHP synthase. J. Mol. Graph. Model. 101, 107735 (2020).
Learn more about how the CSD Ligand Overlay enables drug design as part of CSD-Discovery.
Explore more examples of CCDC tools in action.
Want to try it for yourself? Follow the step-by-step tutorial on Ligand Based Virtual Screening including Ligand Overlay here.