How to Compare the Packing of Your Polymorphs
Polymorphism refers to the phenomenon in which a compound can crystallize into two or more different crystalline forms. This is particularly relevant for pharmaceuticals, as different polymorphs can exhibit different properties, which in turn impact bioavailability and efficacy of the drug.
The study and comparison of polymorphic structures is not always straightforward. The Crystal Packing Similarity and Structure Overlay functionalities available in Mercury can help you find differences and similarities in your polymorphs by exploring the packing environments and conformation.
This blog provides a guide on how to use these functionalities and illustrates various features that you could benefit from when analysing your own polymorphs.
The Family of Polymorphs
Here we will investigate the structures of three polymorphs of a potential tuberculostatic drug that were obtained from different solvents and crystallization methods. These are CSD Entries: DEDMUX, DEDMUX01, and DEDMUX02.
To visualize each structure of the three polymorphs:
- Open Mercury and search for DEDMUX on the “Structure Navigator” bar;
- The CSD Entries of the three polymorphs (DEDMUX, DEDMUX01, and DEDMUX02) will appear on the top of the “Structure Navigator” panel, where you can select each of them and visualize their structure;
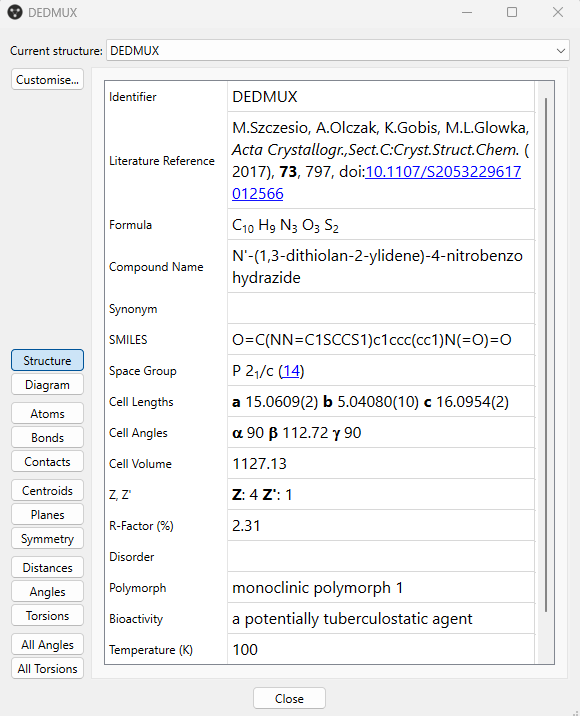
- To find out more information about the selected polymorph you can go to “More Info” > “Structure Information” from the “Display Options” panel, and quickly visualize data such as the unit cell parameters, bioactivity, and details on the type of polymorph (Figure 1, note that the contents of this panel can be customized).
Crystal Packing Similarity
The Crystal Packing Similarity functionality allows the user to compare the packing of polymorphs and understand how similar their solid forms are.
To use the functionality:
- Go to “CSD-Materials” > “Search” > “Crystal Packing Similarity”;
- From the “Packing Similarity Wizard” you can select the structures that you want to compare: for this example, we will add the full family of polymorphs both as Reference and as Comparison Structures;
- In the Reference Structures panel go to “Select” > lookup button > add DEDMUX as Refcode > tick “Enter refcode family” > “OK” > “OK”;
- Once the three structures have been added, do the same for the Comparison Structures panel and then click “Next”;
- Click “Next” two more times if you do not want to change the default options, and then click “Compare”.
Reported in Figure 2 are the results from the Crystal Packing Similarity functionality. A “Searches” panel opens, allowing the user to select the structures to compare, and to visualize them on the Mercury window. In Figure 2, the comparison selected and visualized involves DEDMUX and DEDMUX01, which have 7 out of 15 molecules in common (highest value among the compared structures).
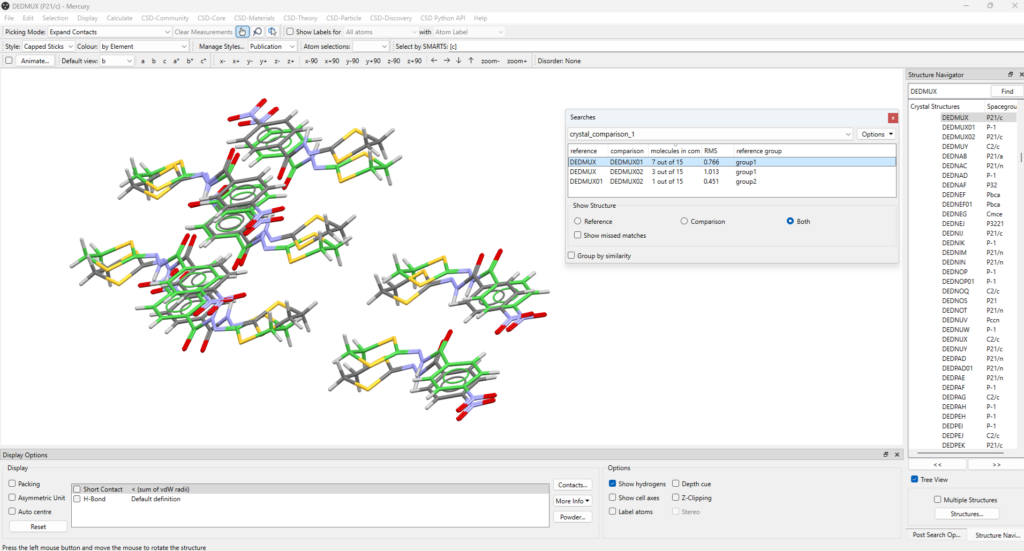
Depending on what you are interested in, at the bottom of the “Searches” panel you can choose to show only the Reference Structure, the Comparison Structure, or both for the specific pair of polymorphs selected. The cell axes can be displayed alongside the packing similarity results by ticking “Show cell axes” in “Display Options”.
You can also select “Show missed matches” to view the molecules in the packing that did not match, which will be highlighted in red.
Finally, you can select “Group by similarity”, a feature that allows the users to quickly compare the features of the polymorphs, such as their space group, density, and unit cell parameters (Figure 3).
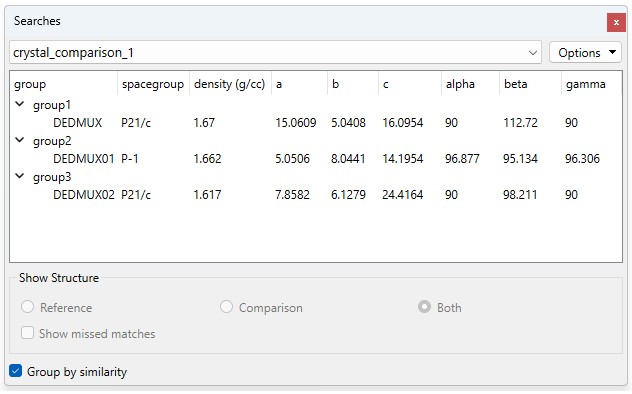
Results From the Crystal Packing Similarity Analysis
The relatively low number of molecules that the polymorphs have in common, and the significant differences in unit cell parameters suggested that their packing is quite different.
By analysing the packing in detail, the conformation seemed to differ the most around the sulphur heterocyclic ring for DEDMUX02 (visualized by selecting the rows involving DEDMUX02 as comparison).
The Structure Overlay function can help you to further understand the differences and similarities here.
Structure Overlay
The Structure Overlay functionality allows the user to overlay two or more structures and to compare their conformations in detail.
To use the functionality:
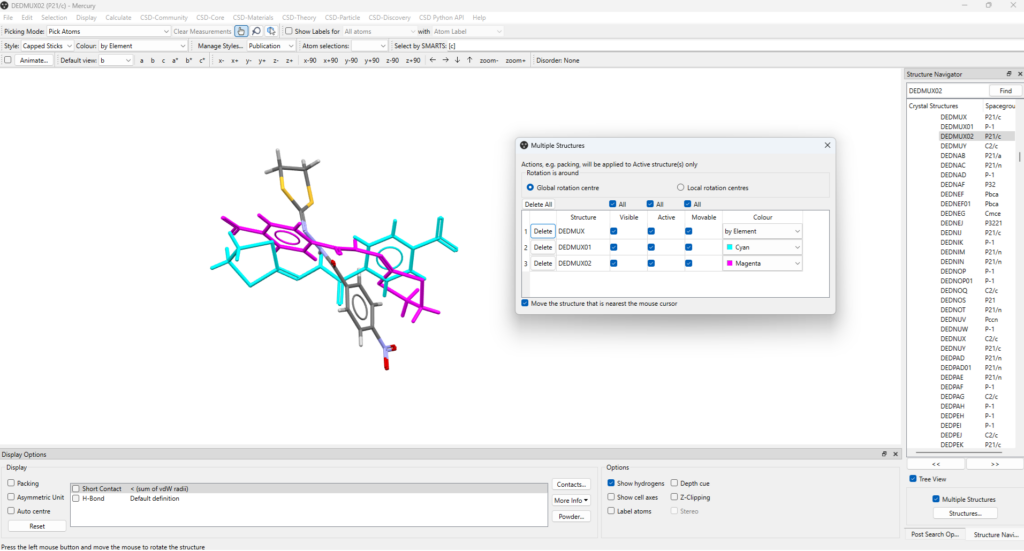
- Tick “Multiple Structures” at the bottom of the “Structure Navigator” panel, then select each CSD Entry you want to compare, one at a time: in this example we use the three polymorphs DEDMUX, DEDMUX01, and DEDMUX02;
- The selected structures will then overlay in the Mercury window, and options that make it easier to distinguish and compare them can be accessed from the “Structures” button at the bottom right hand corner;
- From here, you can change the colour for DEDMUX01 and DEDMUX02 (in the example in Figure 4 to cyan and magenta) and then tick “Move the structure that is nearest the mouse cursor”;
- “Left click + move mouse” will allow you to rotate only the closest of the overlayed structures, “Ctrl + left click + mouse move” will allow you to translate it;
- Orient the structures like in Figure 5 and untick “Move the structure that is nearest the mouse cursor” to avoid moving them further;
- Go to “Calculate” > “Structure Overlay” and you’ll be able to overlay two of the molecules at a time;
- As we were interested in seeing the differences in the conformation of the sulphur heterocyclic rings, the corresponding pair of atoms that we picked for the overlay were on the side of the molecules with the aromatic ring – click one atom in the first molecule followed by the corresponding atom in the second molecule, and repeat until all desired atom pairs are selected (Figure 5);
- After you’ve done this, press “Overlay” to apply to DEDMUX and DEDMUX01;
- To overlay the third structure, delete the pairs of atoms in the “Structure Overlay” panel, untick “Visible” in the Multiple Structures dialog for DEDMUX01, then repeat the process for DEDMUX and DEMUX02.
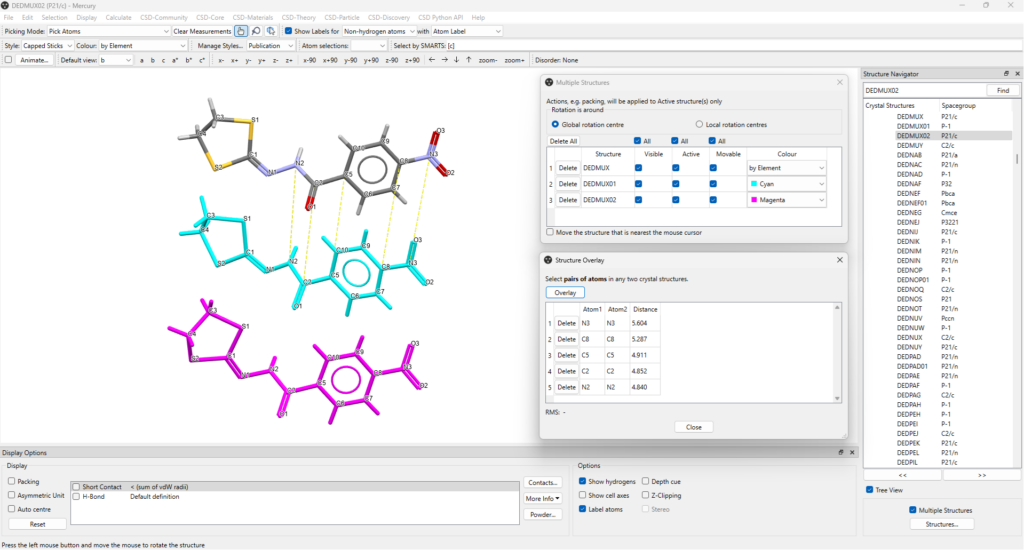
After performing these steps, you should be able to see two of the three polymorphs overlayed.
Make DEDMUX01 visible again from the “Multiple structures” panel to see all the three polymorphs overlayed, as shown in Figure 6. If it is easier, you can go back to visualizing only two of them by making the third one not visible.
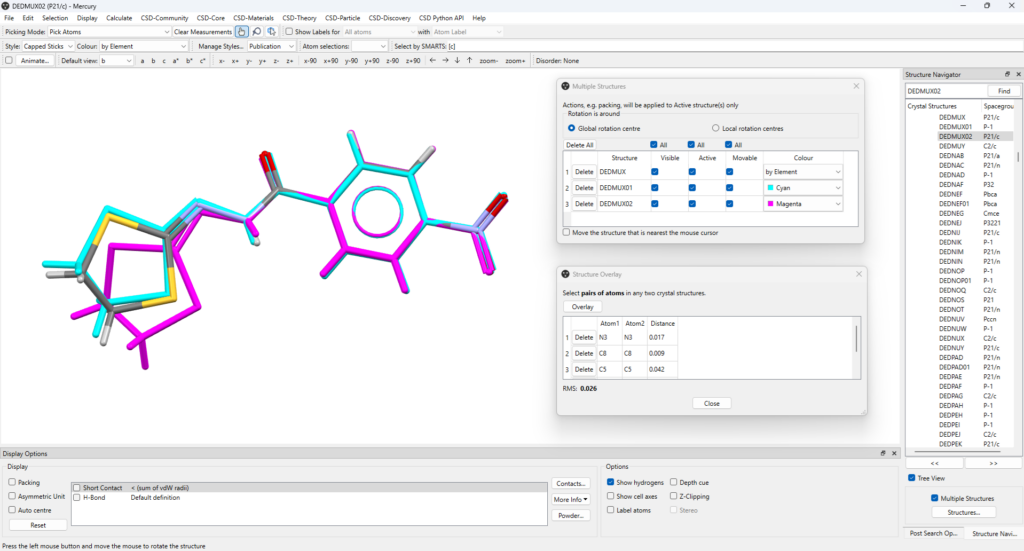
Results From the Structure Overlay Analysis
By analysing the overlay shown in Figure 6, it can be seen that DEDMUX (coloured by element) and DEDMUX01 (coloured in cyan) have a similar conformation, with a very small variation in the sulphur heterocyclic ring.
Instead, the third polymorph, DEDMUX02 (coloured in magenta), differs quite significantly from the other two for the conformation of the sulphur heterocyclic ring.
As the aromatic side of the three molecules overlays almost perfectly, it can be concluded that the main differences in conformations between the polymorphs are mostly dictated by the flexibility on the sulphur heterocyclic ring side of the molecules.
Next Steps
- Discover more about the functionalities available in Mercury to analyse polymorphic structures and assess their stability. Watch our Virtual Workshop: In depth comparison of polymorphic structures using Mercury and find more material, including exercise handouts here.
- Watch the video How to: Overlay structures in Mercury to learn more features available for this functionality and see it in use.
- To discuss further and/or request a demo with one of our scientists, please contact us via this form or .