Getting Started with Protein-Ligand Docking Using GOLD
The protein-ligand docking software GOLD is known for its high accuracy in predicting ligand binding and flexibility in handling diverse protein docking scenarios relevant for drug discovery.
In this blog, we provide an introduction to GOLD to get novice users started, and do a deeper dive for the more experienced docking scientist.
Protein-Ligand Docking with GOLD – Terminology
We will start by defining some key terms in this field:
- Proteins are large biomolecules and macromolecules that comprise of one or more long chains of amino acids.
- Ligands are small molecules that bind to the protein and can change the protein function.
- Functional waters are found in the binding site of a protein and mediate the interactions between the ligand and the protein.
- Docking studies are computational techniques to explore possible binding modes of a substrate to a given receptor, enzyme or other binding site.
- A binding site is a specific region (or atom) in a molecular entity that is capable of entering into a stabilizing interaction with another molecular entity.
- Scoring functions are mathematical functions used to approximately predict the binding affinity between two molecules after they have been docked.
- GOLD stands for Genetic Optimization for Ligand Docking. It is a software based on a genetic algorithm, for docking flexible ligands into protein binding sites.
Find more information in our glossary included in the self-guided workshop “Introduction to protein-ligand docking with GOLD“
Where Can GOLD Be Helpful?
Like all the components of the CSD-Discovery suite, GOLD is a knowledge-based tool for drug design. This means that it uses the structural information stored in the Cambridge Structural Database (CSD) to make reliable predictions and empower molecular discovery in different steps of the drug design pipeline.
GOLD has proven success in virtual screening, lead optimisation, and identifying the correct binding mode of active molecules, and has been validated in the following peer-reviewed publications:
- A new test set for validating predictions of protein-ligand interaction, Nissink et al., Proteins, https://doi.org/10.1002/prot.10232
- Improved protein–ligand docking using GOLD, Verdonk et al., Proteins, https://doi.org/10.1002/prot.10465
- Diverse, high-quality test set for the validation of protein-ligand docking performance, Hartshorn et al., J. Med. Chem., https://doi.org/10.1021/jm061277y
- Pose prediction and virtual screening performance of GOLD scoring functions in a standardized test, Liebeschuetz et al., J. Comput. Aided Mol. Des., https://doi.org/10.1007/s10822-012-9551-4
With GOLD it is possible to perform protein-ligand docking, pose prediction, run docking experiments with water molecules, and choose between multiple scoring functions. Ensemble docking can also be performed, which allows one molecule to be docked in multiple proteins, as well as covalent docking. Several types of constraints can be applied to your docking experiments, and all this can be done on a personal computer, on a cluster, or on the cloud.
Steps in Molecular Docking with GOLD
The first step to perform molecular docking is the target selection, which can be for example a co-crystal structure of a protein-ligand complex from the Protein Data Bank (PDB). Once this has been opened or imported in GOLD, the software allows users to prepare the protein for docking by:
- Adding missing hydrogens
- Checking the protonation of residues
- Removing waters or retaining the required ones
- Extracting the ligand structure and saving it
GOLD can then be used to define the binding site, and there are four ways to do this, depending on what you want to explore and on whether a co-crystal structure of a protein-ligand complex is available or not. More information can be found in paragraph 4.6.2 Defining a Binding Site from an Atom of our “GOLD User Guide” available from the GOLD Documentation & Resources page.
With GOLD you can then select one or more ligands for docking, and choose the preferred scoring function between four different ones:
- GoldScore
- ChemScore
- ASP – the Astex Statistical Potential
- ChemPLP
The docking calculations are then ready to be set up and run with GOLD. The chosen ligand will dock into the defined binding site, and the results of the calculations will be accessible for further analysis. This includes allowing the users to look at the fitness scores and RMSDs obtained from the calculations, but also to visualize and measure the interactions formed. More information can be found in paragraph 16 Viewing and Analysing Results of our “GOLD User Guide“.
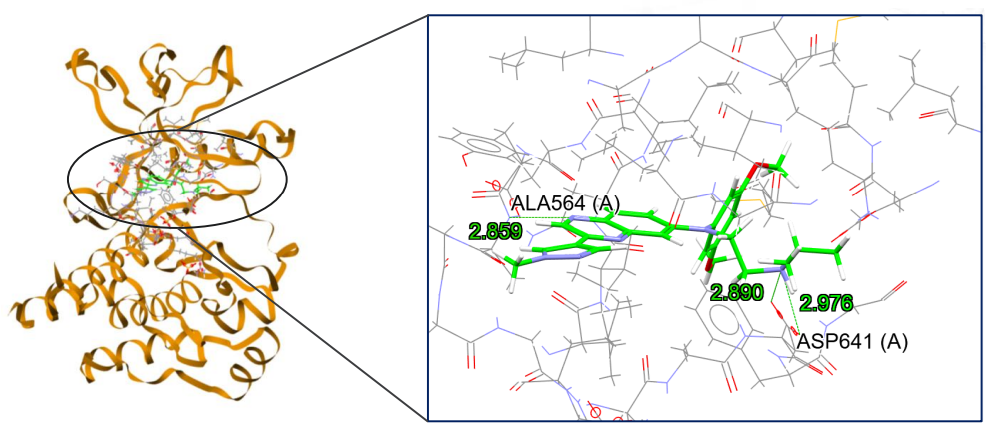
Using GOLD Through the Hermes Interface
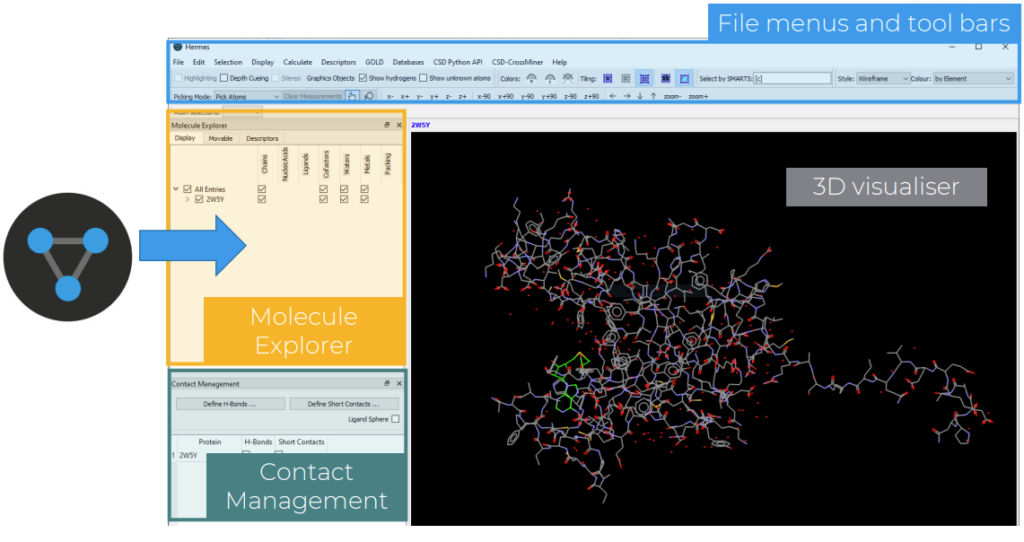
GOLD can be accessed through the Hermes desktop interface.
When you open Hermes, a variety of panels will be available:
- The 3D visualizer, which populates with the crystal structure of the selected protein-ligand complex when this is opened or imported
- The Molecule Explorer panel, which lists the different components of the crystal structure, such as chains, ligands, waters in the protein, and allows the user to show only those of interest and change their colour or style
- The Contact Management panel, useful to show interactions in the structure such as the hydrogen bonds
- File menus and tool bars, to access different functionalities and different visualization options
To load a structure the user can go to “File” > “Open” or import it from the PDB through “CSD Python API” > “Import” > “fetch_from_pdb.py”.
A variety of visualization options are also available under “Display”, including the “Ribbon & Tubes” option and the possibility to choose different colours to display the protein.
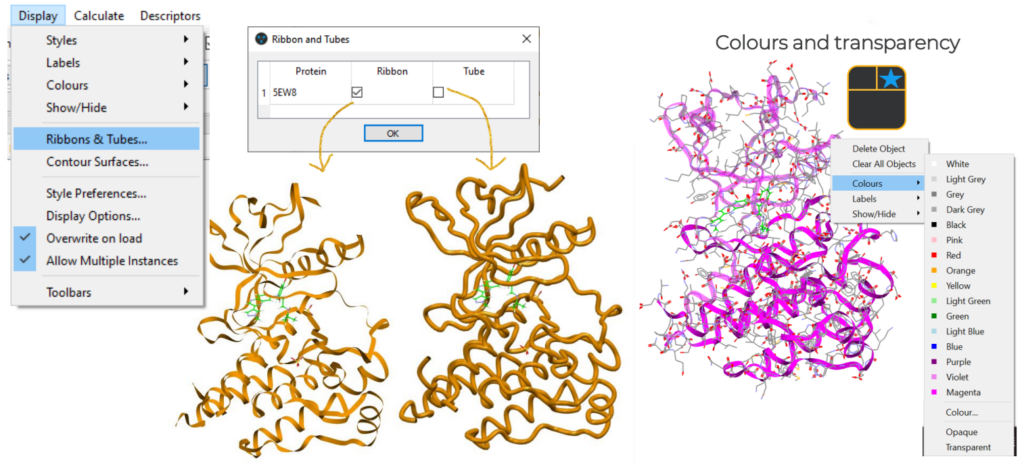
Finally, GOLD can be accessed from the menu on the top of the Hermes interface by clicking “GOLD” > “Wizard”, where the docking study can be set up and started.
Using GOLD – Where to Start
The CCDC provides a series of workshops and short videos that can help you getting started with GOLD.
The self-guided workshop “Introduction to protein-ligand docking with GOLD” shows how to perform protein-ligand docking of a Kinase inhibitor. You’ll find a step-by-step guide on how to:
- Import a protein co-crystal from the PDB
- Prepare the protein crystal structure for docking
- Perform a molecular docking experiment in GOLD
- Analyse the results obtained from the docking experiment
More training material can be found on the CSD-Discovery Workshop page, where a list of self-guided workshops is available on more specific topics such as how to perform ensemble docking, pose prediction, and docking with waters in GOLD.
A video playlist of tutorials and tips for GOLD is also available on our YouTube channel.
Next Steps
There are many case studies in our blog showing how GOLD has been used with different modalities and targets. Here are a few white papers with greater detail of specific studies:
- Protein–Ligand Docking: AlphaFold2-predicted vs. X-ray Determined Protein Structures
- GOLD Study: Docking Small Molecules to Nucleic Acid Targets
- GOLD Docking Study: Linear Peptide Docking
To discuss further and/or request a demo with one of our scientists, please contact us via this form or .