CSD Data Curation – The Human Touch
Rather than tell you how long I have worked at CCDC as a Scientific Editor in the Database Group, perhaps I will instead measure the time in the growth of the Cambridge Structural Database (CSD) and tell you that when I started working here, the CSD had just passed the 180,000 mark. As I’m sure you know, today, more than 800,000 structures later, we are very close to entering the 1 millionth structure into the CSD.
But what do we mean when we say ‘entering’ data into the CSD, a process we refer to as data curation?
Update 05/05/2021: Since the writing of this blog, the CSD has surpassed 1.1 million unique crystal structures.
Digital transformation and data curation
Although the way in which data is deposited has changed significantly over this period, it may surprise you to know how much of a human touch is still involved with the production of the CSD. Back in those days of 180,000 (still not telling you when!) CSD entries, just over half of structures were sent to us in CIF format. Nowadays, this number is well over 99%. This might lead you to think that there would be less need for human intervention in producing the CSD. However, as you can see below, the Database Group here at CCDC is still quite sizeable with a significant portion of our time being spent on building and maintaining the CSD.

The CCDC Database Team – January 2019
In the days when almost half of the data was arriving at CCDC in non-CIF format (and in many cases hard copy of one type or another), a significant amount of human effort was required to convert the hard-copy data we received into an electronic format, with the inevitable introduction of a certain amount of human error. Thankfully, the software we used in those days was well set up to perform internal checks on atomic coordinates versus reported bond lengths and allowed such errors to be corrected. A considerable amount of human effort was also required to construct chemical diagrams and to produce chemical names.
These days almost all data is deposited electronically in CIF format and with the introduction almost 6 years ago of CSD-Xpedite [1], the process of deposition of data at the CCDC and the initial processing of CIFs is automated for the majority of structures. CSD-Xpedite includes a program known as DeCIFer [2] which uses a Bayesian approach to suggest a likely chemical representation based on a combination of the observed geometry of molecules in a structure and knowledge based on the hundreds of thousands of (editor-validated) structures already in the CSD. DeCIFer also attempts to resolve disorder based on the reported atom occupancies in a CIF. We also have software which attempts to generate a chemical name automatically using third party software [3].
Adding the human touch
Although the data output from DeCIFer gives us a good starting point, every one of the (typically 50,000+ per year) new structures entering the CSD is manually checked by one of the Scientific Editors. In the case of many small organic molecules, little intervention is required beyond a check for chemical sense, although human error in the generation of a structure still cannot be entirely overlooked. For instance, if hydrogen atoms have not been located and/or are mis-assigned then the algorithm will struggle and a human eye is much more suited to spotting the problem.
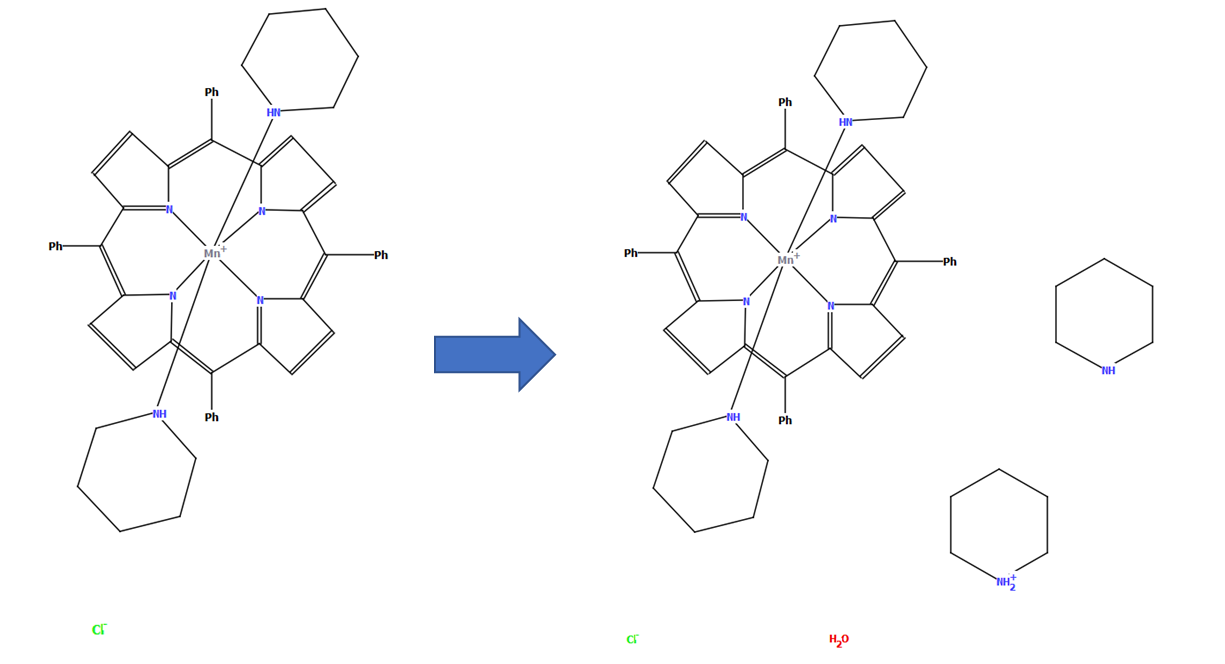
Stepping up one level of complexity to small organometallic compounds; a further internal check on the consistency of data is provided by manual checking of metal oxidation states and charge balance, with reference to the publication if necessary. A human eye is particularly important for tricky cases such as redox-active ligands, radicals and metals with multiple possible oxidation states.
A great deal of human effort is required in larger organometallic complexes in which, for very good reasons, the assignment of hydrogen atoms might not be possible. At this stage, we as Editors refer to the associated publication and do our best to represent in the CSD chemical diagram, the chemistry described in the paper, whilst also trying to maintain a level of consistency in representation throughout the CSD. The amount of void space is calculated for every structure and any mention of PLATON/SQUEEZE or OLEX2/MASK is picked up either automatically or by manual checking of the publication and its supporting information. As well as looking for information to include in the CSD, this can of course sometimes be a case of checking all sources to ensure that information is not present! The reported formula and stoichiometry are manually checked for every structure since this provides a powerful way of spotting numerous potential problems.
Basic automation versus manual and automated curation
Ensuring that each entry is represented in a readable and understandable way, as well as in a way that ensures that data can be reliably searched and used in downstream data analysis to gain new insights, can be a complex process. Below are a few examples of how a structure might look using basic automation compared with how we represent it in the CSD using both automated and manual processes to ensure that the structures have the correct chemical connectivity and that the chemistry is displayed in a meaningful way.
CSD refcode TIZGIV with a diagram as it might appear in projection from the atomic coordinates (left), a diagram generated by DeCIFer (with disordered solvent) (middle) and in its finished form with the solvent molecule drawn correctly and the Co-Co bond removed as noted in the description of the structure in the associated publication (right)
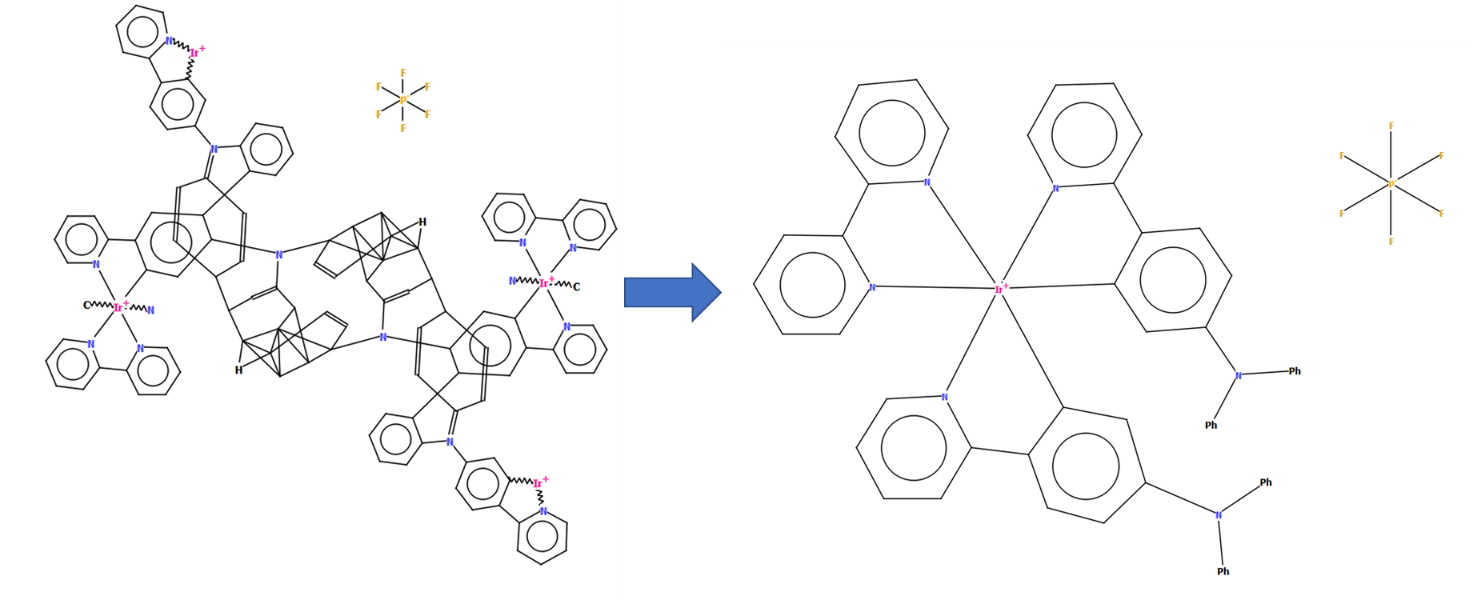
CSD refcode NIWBIH with a diagram generated by DeCIFer showing disorder (left) and in its finished manually-edited form with a clearer representation of the structure in a chemical diagram without disorder (right)
CSD refcode TOBTOW with a diagram generated automatically by DeCIFer (left) and the finished diagram complete with additional species treated using PLATON/SQUEEZE and described in detail in the paper (right)
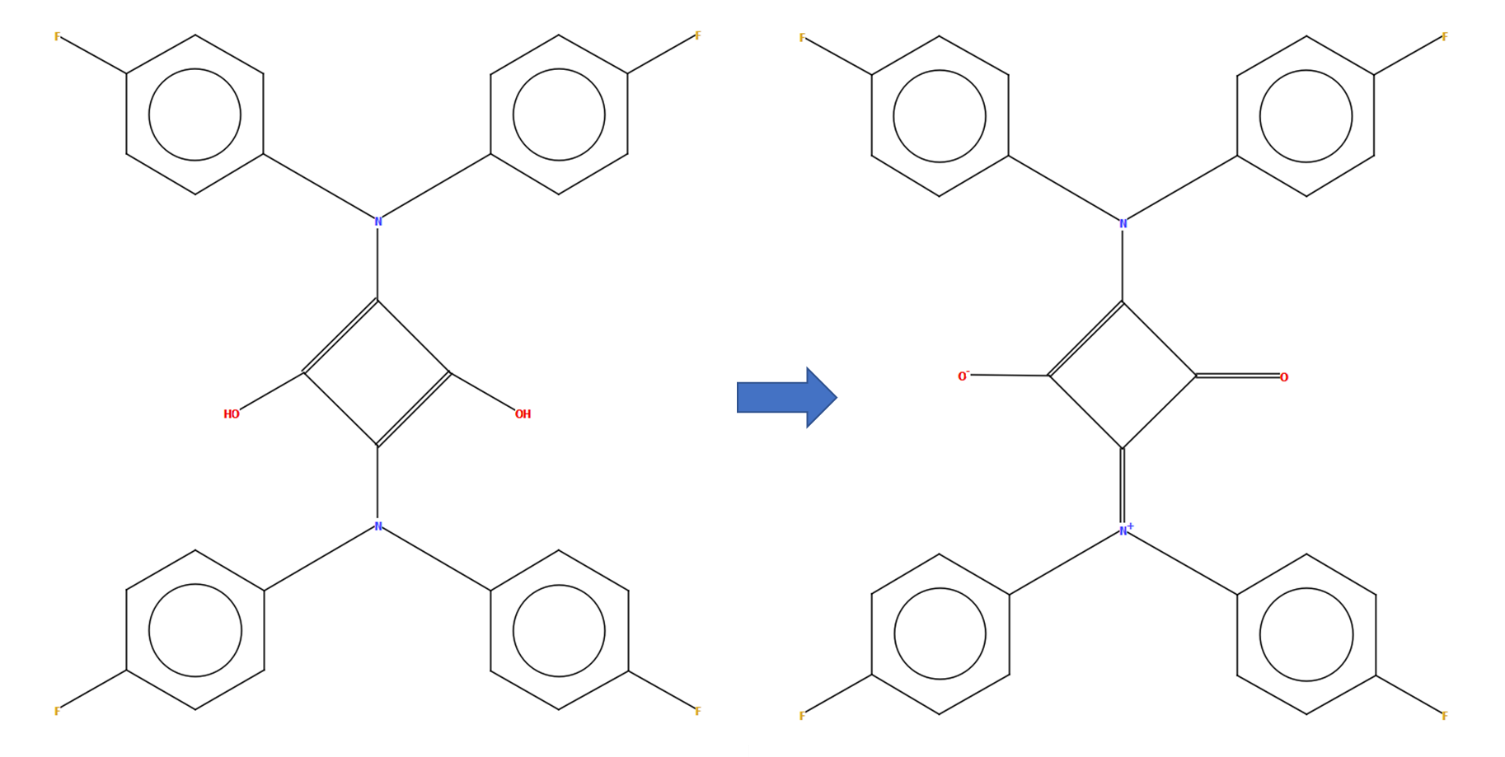
CSD refcode LIXDAA with a diagram generated automatically by DeCIFer (left) and the manually edited diagram representing the more complex charge-separation found by reading the associated publication (right)
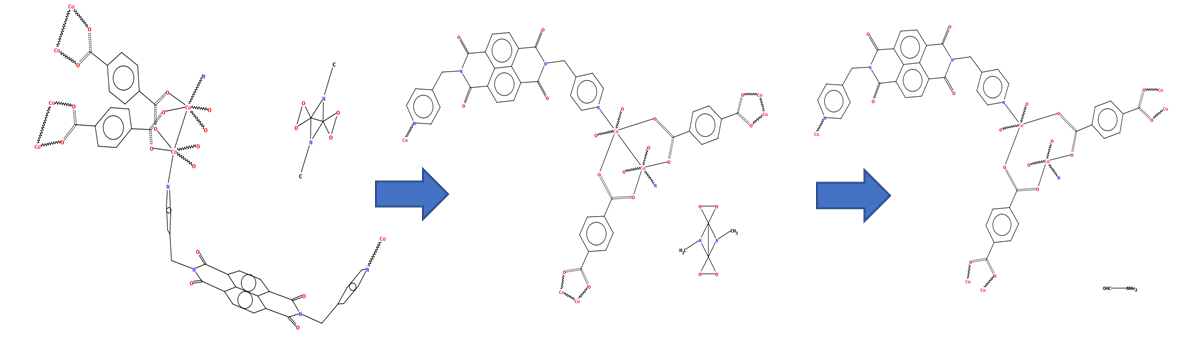
The example below of a polymeric copper oxo-molybdate structure [CSD refcode TISRUL] illustrates a way in which human intervention makes a structure more accessible to a human CSD-user. The algorithm chose to split the structure in the middle of the molybdate unit, resulting in a diagram which at first glance is difficult to understand, whereas the second part of the figure shows the finished version with intact oxo-molybdate clusters.
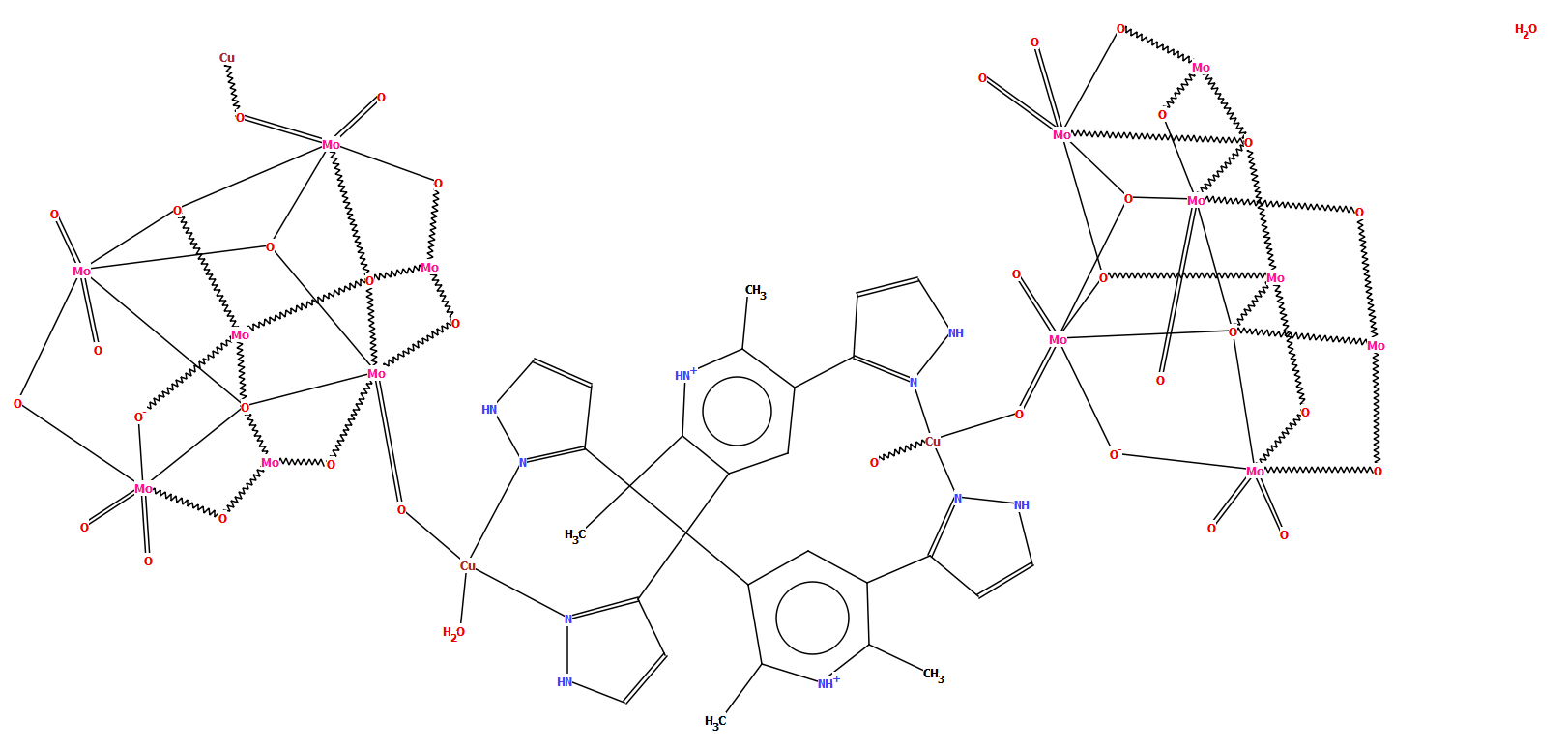
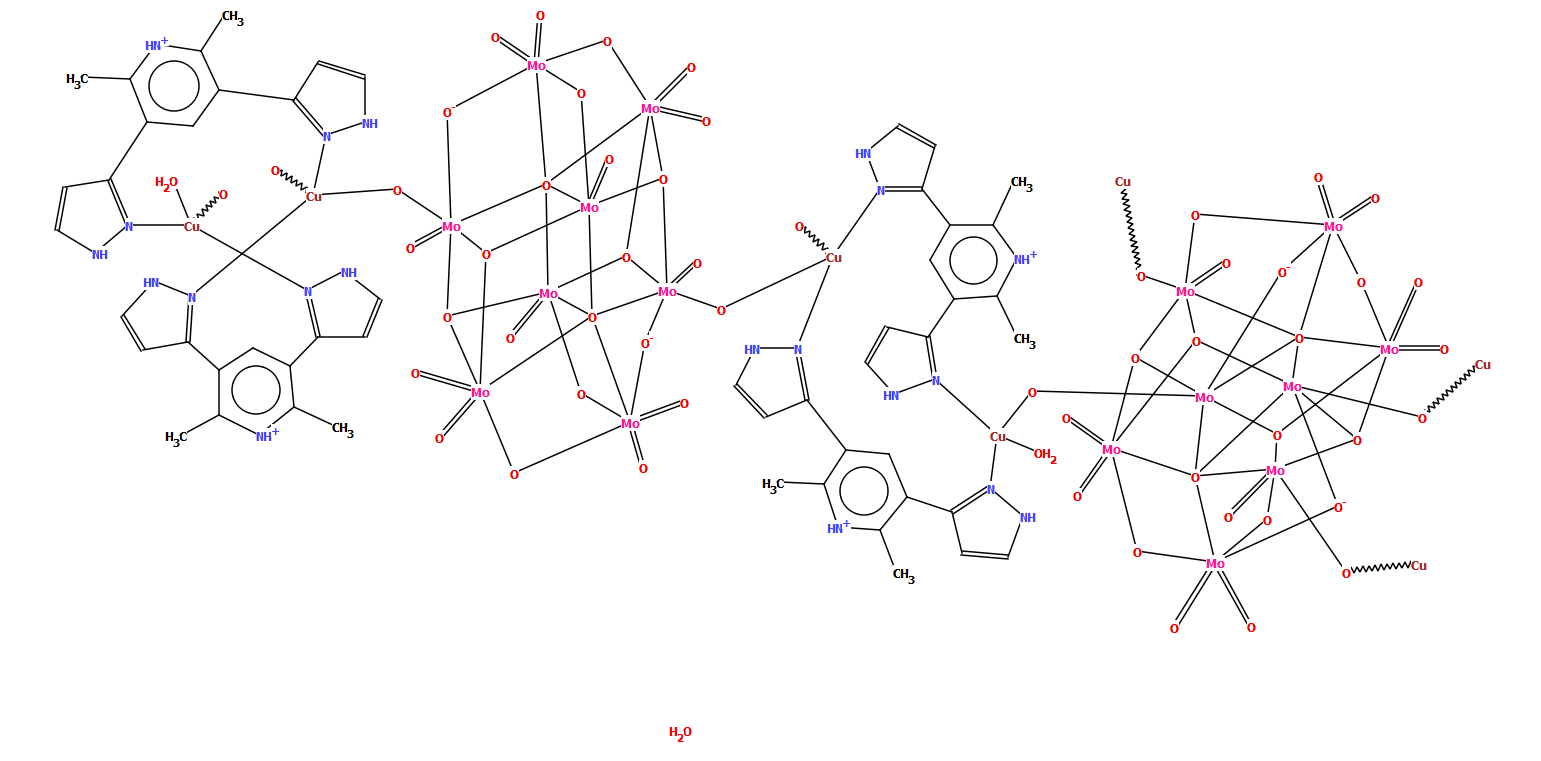
CSD refcode TISRUL with a diagram generated automatically by DeCIFer and the finished diagram
It is not just these obvious issues that are spotted. Sometimes the care and attention in curating data into the CSD leads to us spotting far more subtle issues, which can lead to us corresponding with depositors and as a result receiving revised data which improves the quality of the CSD and ultimately benefits the community.
As crystallographic techniques have improved, structures of greater and greater complexity are solved, which can be quite challenging to understand and represent. An example of this is MOF structures, for which the determination of the overall framework structure might be sufficient for the purpose of the science involved, but in trying to produce a CSD entry we assimilate and summarise a significant amount of complex information into a single chemical diagram. Trying to represent even a relatively simple MOF structure in a single chemical diagram can be complicated.
Maintaining consistency to improve discoverability
There are sometimes multiple different representations and interpretation in a publication and often different research groups can interpret the chemistry of the structure in different ways. Therefore, one of our roles is to try our best to maintain a level of consistency of representation in the CSD and to make structures as discoverable as possible.
Discoverability is also in our minds when producing chemical names. Automatically generated names [3] can be a useful starting point as we try to follow IUPAC naming conventions where possible and appropriate, but we also work on making names more human-friendly as illustrated by the relatively simple example of CSD refcode TIVQEX.
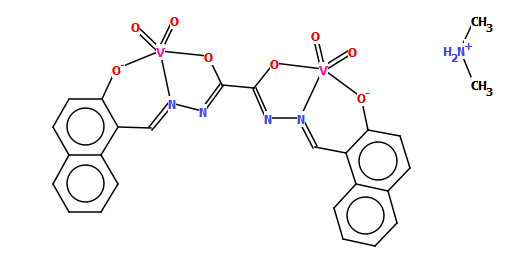
The name generated by software for this compound was:
bis(dimethylammonium) {mu-[N$1!-{[2-(hydroxy)naphthalen-1-yl]methylidene}-N$2!-{[2-(hydroxy)naphthalen-1-yl]methylidene}ethanedihydrazonato]}-(tetraoxo)-di-vanadate
Which with a few improvements becomes:
bis(dimethylammonium) {mu-N$1!,N$2!-bis{[2-oxynaphthalen-1-yl]methylidene}ethanedihydrazonato}-tetraoxo-di-vanadium(v)
Software is used to guide us towards structures which are possibly the same, and therefore should be grouped into the same refcode family, although a Scientific Editor makes the final decision on this, taking into account such matters as polymorphism, stereochemistry etc. The process of assigning structures to Chemical classes (Carbohydrates, amino-acids, porphyrins, steroids etc) also takes some manual effort.
I find it interesting that although our focus as Scientific Editors has changed over the years, helped by great improvements in our database-building software, due to factors including the solution of increasingly complex structures by crystallography, our part in producing the CSD is just as important as it ever was. Hopefully the human touch in the construction of the CSD has a great many benefits but on those rare occasions when you spot a potential problem with a CSD entry, please feel free to contact us at and we will look into the matter. We treat the CSD as a continuously evolving database and annually we undertake projects to enhance and improve existing CSD entries. This will be the subject of another blog article in the very near future.
[3] ACD Name – https://www.acdlabs.com/