Covalent Docking Enhancements in GOLD and Ligand Overlay in the CSD Python API
Covalent docking in the protein–ligand docking software GOLD is now faster and easier than before, and you can now overlay ligands in a repeatable manner with the CSD Python API.
Covalent Docking Enhancements
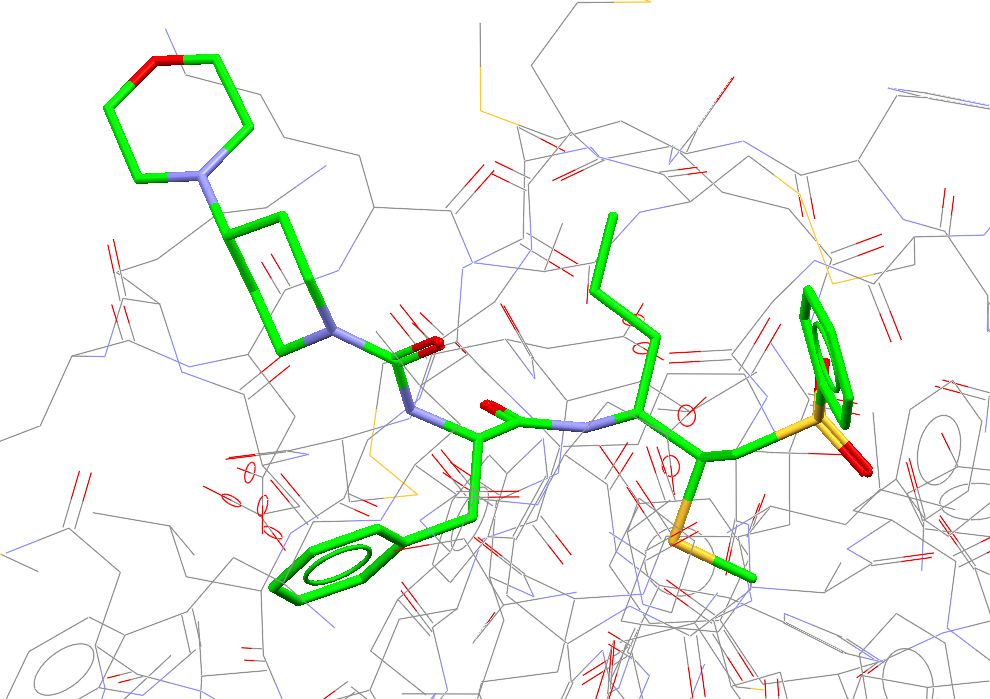
Traditional molecular docking predicts non-covalent interactions like hydrogen bonds and hydrophobic contacts. Covalent docking goes a step further — it models the formation of a new chemical bond between the ligand and the protein during binding. This is crucial for studying irreversible inhibitors, often used to treat cancer, infectious diseases, and immune disorders.
GOLD now automatically:
- Detects reactive groups (warheads) on ligands.
- Matches them with compatible residues.
- Provides users with a choice of reaction transformation.
- Performs full chemical transformations: for example, breaking open rings, adjusting geometry, and forming new bonds.
Instead of merging the protein and ligand into a single molecule, GOLD extends the ligand to include the side chain of the reactive residue. The protein is “trimmed” by replacing the side chain with an alanine, making everything clean and consistent.
This design has several advantages:
- Better stability for the covalent bond during docking.
- Easier handling of different stereoisomers and tautomers.
- Clearer control for users selecting reaction options.
Any new covalent docking features in GOLD are also available through the CSD Python API.
Ligand Overlay Now in the CSD Python API
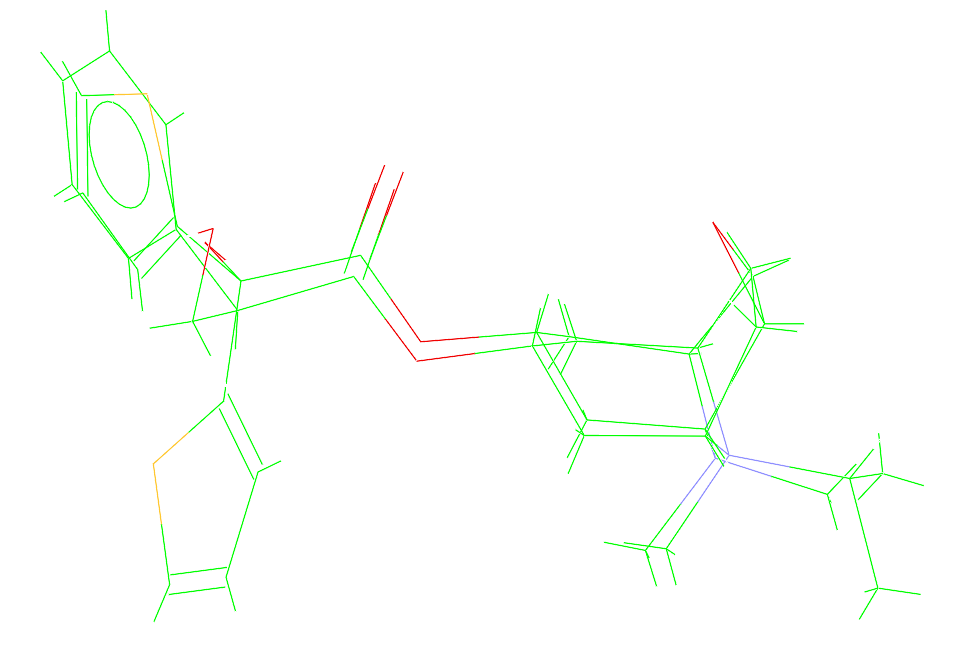
Ligand Overlay is now available through the CSD Python API.
Overlaying ligands is crucial for:
- Pharmacophore modeling.
- Comparing docked poses.
- Ligand-based drug design and lead optimization.
What’s New?
- Easier integration into larger automated workflows.
- Better stability: fixed GUI bugs that may have previously caused crashes.
- Enhanced control: inspect overlay solutions (such as volume and RMSD) directly in scripts.
Summary
| Feature | Benefit |
|---|---|
| Covalent Docking Enhancements | Model complex covalent reactions automatically. |
| Warhead Detection | Easy and flexible setup for users. |
| CSD Python API Ligand Overlay | Automate pharmacophore and overlay workflows. |
| GUI Improvements | Cleaner, more intuitive experience. |
These updates make GOLD much more powerful for users working on irreversible inhibitors, fragment-based design, and ligand-based drug discovery. With covalent docking and ligand overlay now seamlessly scriptable in Python, it’s easier than ever to integrate GOLD into modern computational pipelines.
Next Steps
To discuss further and/or request a demo with one of our scientists please contact us via this form or .
More information on our data and software for drug discovery.
Check out the Cambridge Structural Database (CSD) for yourself and see how your research can benefit from the combined knowledge of over 1.3M small-molecule organic and metal-organic crystal structure data.
Proprietary data? Our team can curate your proprietary data into a Cambridge Structural Database (CSD)-like database, accessible through a simple browser-based interface. This is all done within your firewall to comply with your data security requirements.
More information on CSD software trusted by academic and industrial institutions around the world.
See case studies of the CSD in action, driving forward the boundaries of scientific research.