2019 CSD Release: A New Beginning
This release marks the beginning of a new chapter in the history of the Cambridge Structural Database (CSD). We have just completed a multi-year software development project to replace the search engine at the heart of our well-known search program ConQuest. This not only brings into alignment the search functionality behind ConQuest, Mercury, WebCSD and the CSD Python API, but it enables much more flexible, dynamic and advanced searching of the CSD in the future.
Completely rewritten C++ search engine powering ConQuest which now uses consistent, shared methodologies with Mercury, WebCSD and the CSD Python API.
The Fortran search engine behind ConQuest, amusingly named “Thomas the Search Engine” or “Thomas” for short, dates back to the 1970s and 1980s, when the CCDC was first developing electronic search capabilities. This has really stood the test of time, continuing to be effective for many years. The first steps towards a new, more modern search engine were taken in the early days of our visualisation program Mercury (in the 2000s) through writing new, reusable C++ chemistry algorithms. The introduction of new, faster 3D search options in Mercury 2.0 in 2008, followed by further new search capabilities in WebCSD in 2010, continued this movement by implementing C++ search functions in parallel to ConQuest. This release completes the transition.
The evolution of our entire software portfolio continues at pace, and is now significantly aided by this step forward to unify all our searching methodologies. Some of the immediate benefits in ConQuest this year include many complex substructure searches becoming faster, as well as text, element and formula searches becoming more effective due to the more modern algorithms being used. The search engine, Thomas, and the format of the CSD itself have been inextricably linked in the past, thus restricting our development options. This is no longer the case and we will be able to introduce more data types and search options in the future. One example is the expansion of CSD entries displaying anisotropic displacement parameters (ADPs) in Mercury from just over 220,000 last year to over 675,000 structures this year! You should expect to see more expansion of data fields and search types in the coming years, so watch out for future communications from us seeking your input on this.
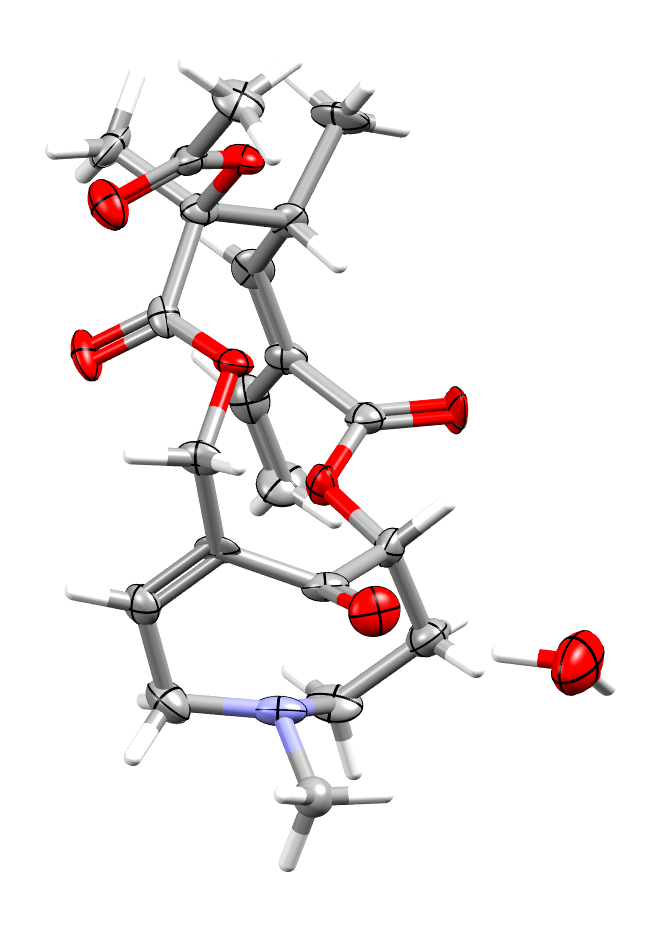
The structure of the alkaloid clivorine illustrating the Bürgi-Dunitz angle, with ADPs displayed at the 80% probability level (refcode: CLIVOR11, DOI: 10.5517/ccdc.csd.cc16txtr)
Of course, this isn’t the only significant change in the CSD release this year – we have also been working hard in other areas. We have been extending CSD-CrossMiner to better bring together chemistry and biology structural data in the CSD and PDB and expanding the capabilities in the CSD Python API to enable better connectivity for you, amongst other things. To read more about these changes, see our 2019 CSD Release What’s New guide. The CSD also continues to grow and evolve – to find out more about the over 57,000 new entries and over 80,000 improvements to existing entries, see our 2019 CSD Data page.
As always, let us know what you think about the latest release and what improvements you would like to see in the future through , or any of our CCDC social media channels (Facebook, Twitter and LinkedIn). Any comments or questions from our user community are always welcome!
Pete Wood, CSD-System and CSD-Materials Product Manager
Tags