WebCSD v2 developing even further
We are excited to announce a range of new features that have been introduced to WebCSD v2 – unit cell search, similarity search and query highlighting.
Our web-enabled CSD structure search capabilities are expanding to include similarity searching. The structure search tab within WebCSD v2 now includes three different search settings – exact, substructure and similarity. This means that you can use this application to search for exact molecules, fragments within molecules or similar molecules. The molecular similarity option may be less familiar to some users, but essentially this works using molecular fingerprints. The drawn query molecule is analysed based on atom types, bond types and bonded paths through the molecule to build up a fingerprint and the database is then searched for molecules with closely matching fingerprints. Database hits will have a similarity score between 0 and 1, with 1 being identical and 0 being completely dissimilar, though the search has a cut-off of 0.7 for the similarity score. This search type is great for finding closely related analogues to a given target molecule.
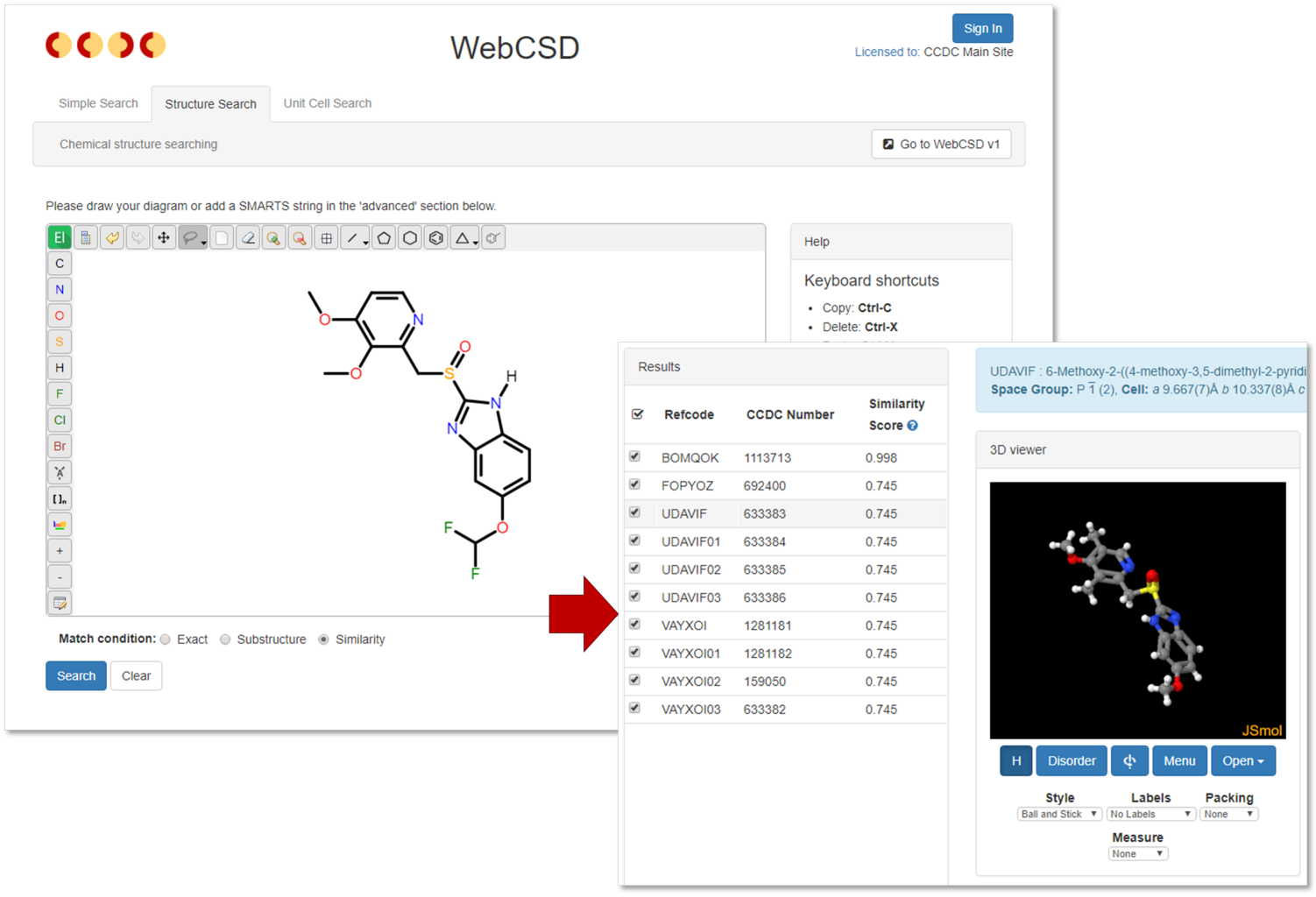
The structure search functionality now provides a third option – similarity – this will find hits similar to the drawn query molecule based on molecular fingerprints.
The other major new search type being introduced to the WebCSD v2 interface is the unit cell search option. This is primarily designed to allow easy searching of the CSD using pre-experiment crystal unit cell details before collecting a full diffraction data set. Unit cell searching allows quick confirmation that known crystal samples are the same as previously published cell dimensions, that new crystal samples have not been previously published and also that new crystal samples are not starting materials or a reaction by-product that has been crystallised by accident. The search uses a reduced cell search methodology that is the same as that used in CellCheckCSD and will be introduced in ConQuest in the 2018 CSD Release.
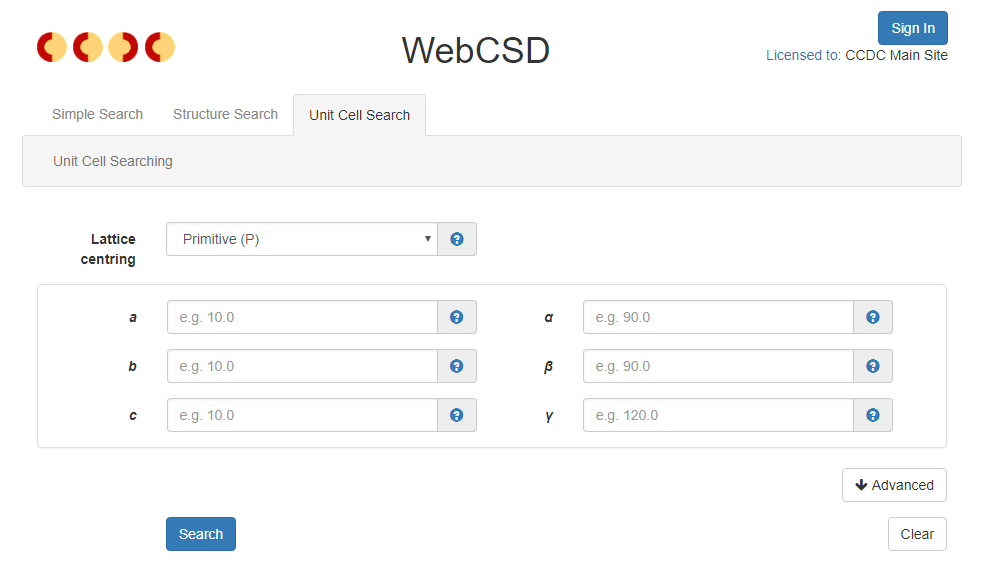
The new Unit Cell Search provides easy access to reduced cell searches against the CSD.
Our text/numeric and structure search options will also now provide highlighting of the query in the results. This makes it a lot easier to see quickly why a given entry has been matched in the search results.
As many of you know, version 1 of WebCSD relied heavily on the use of Java applets for visualising structures, sketching and for browsing search results. With the addition of the new search and highlighting features mentioned above, our new JavaScript-based system WebCSD v2 will now be fully replacing WebCSD v1 over the next few weeks. Do let us know if you have any questions or comments about this transition. WebCSD v1 will remain available for a little while longer as a legacy system and you will be able to access it via a button in the WebCSD v2 Structure Search page (“Go to WebCSD v1”).
We hope that you are finding WebCSD v2 a pleasure to use and that the new features we have added are helpful. Please do let us know what you think about the software and what features you’d like to see in the future – all feedback is welcome via . As with our other applications, more information is also available about WebCSD though our online WebCSD FAQs and our WebCSD product page.
Pete Wood, Product Manager for the CSD-System