2017 CSD Release: Better than Ever
The 2017 CSD Release is now available for download from the CCDC website – if you haven’t already installed it, then hopefully this post will persuade you to do so right away! There is a whole range of improvements this year, both within the database itself and the software, but the overarching focus has been on improving the usability of the system based on feedback from you – our user community.
As always, the CSD release this year contains an even greater number of new entries – the database (with the November 2016 update included) now contains a staggering 858,655 entries. We have also made a big effort this year to improve and enrich existing entries in the CSD, resulting in targeted enhancements to a record-breaking 73,985 existing CSD entries. The improvements in the last twelve months include adding cross-references from entries to DrugBank and the PDB, adding more article DOIs, and augmenting over 21,000 entries with validated metal oxidation states.
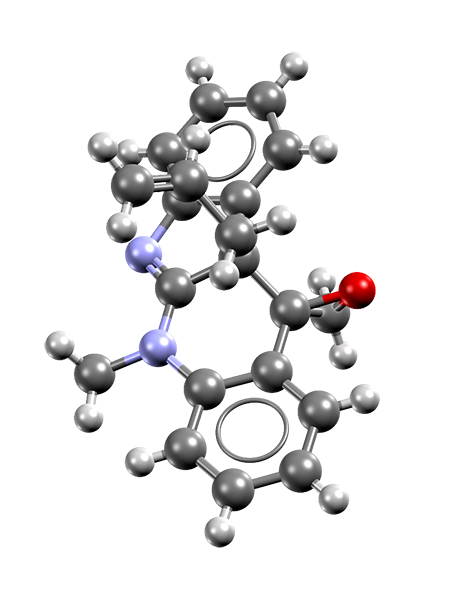
The 850,000th entry in the CSD, an intermediate in the total synthesis of dehaloperophoramidine (refcode: USOPEZ, DOI: 10.5517/ccdc.csd.cc1lm4cq)
One of the most significant improvements we’ve made to the CSD software for this release is to provide a self-contained Python package that includes the CSD Python API and is set up behind the scenes with every installation. This means that all users will be able to immediately make use of the custom Python-enabled menu options that we’ve provided within Mercury. Users can also add their own tailored functionality to Mercury or produce command-line Python scripts even more easily than before.
In CSD-Discovery we’ve made significant improvements to the usability of our leading docking package GOLD. We are providing the first implementation of docking within the CSD Python API and are streamlining our licencing functionality for use on computational clusters. In addition, we now have a complete ligand-based virtual screening workflow to complement the structure-based design capabilities of GOLD. This ligand-based approach includes generation of experimentally realistic conformer ensembles, construction of pharmacophores using our improved Ligand Overlay application and screening with the new Field-Based Ligand Screener application within the CSD Python API.
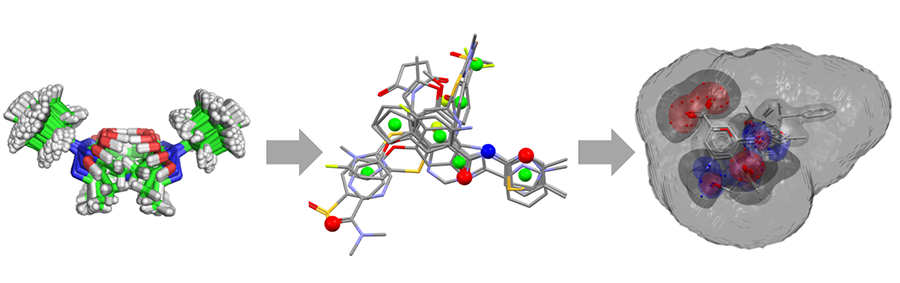
Ligand-based virtual screening workflow in CSD-Discovery
We’ve also been focussing on usability within CSD-Materials this year. Both the Crystal Packing Similarity and Hydrogen Bond Propensity applications have been enhanced to make them simpler and more intuitive for users. When comparing structures, it’s now much easier to pick out what is different, as well as what is similar. The process for calculating hydrogen-bond likelihoods has also been significantly overhauled to streamline and simplify it. Of course, there is much more to come in this area, so do continue to tell us about your experiences using the features.
We hope that you enjoy using the latest release of the CSD this year. If you do not already have access to CSD software at your organisation and are interested, please get in touch via . If you have any feedback about the 2017 CSD Release, or would like to suggest improvements that we could make in the future, please do contact us through . We are always happy to hear from our user community!
Pete Wood, Product Manager for the CSD-System